In this workflow, differential gene expression analysis will be performed on host-transcriptomics data.
First, install all required packages.
# check if BioCmanager libraries are already installed > otherwise install it
if(!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager",repos = "http://cran.us.r-project.org")
if(!"rstudioapi" %in% installed.packages()) BiocManager::install("rstudioapi")
if(!"baySeq" %in% installed.packages()) BiocManager::install("baySeq")
if(!"DESeq2" %in% installed.packages()) BiocManager::install("DESeq2")
if(!"edgeR" %in% installed.packages()) BiocManager::install("edgeR")
if(!"bioDist" %in% installed.packages()) BiocManager::install("bioDist")
if(!"biomaRt" %in% installed.packages()) BiocManager::install("biomaRt")
if(!"dplyr" %in% installed.packages()) BiocManager::install("dplyr")
if(!"magrittr" %in% installed.packages()) BiocManager::install("magrittr")
if(!"EnhancedVolcano" %in% installed.packages()) BiocManager::install("EnhancedVolcano")
#Regular R packages:
if(!"ggplot2" %in% installed.packages()){install.packages("ggplot2")}
if(!"limma" %in% installed.packages()){install.packages("limma")}
#if(!"R2HTML" %in% installed.packages()){install.packages("R2HTML")}
#load packages
library(rstudioapi)
library(baySeq)
library(DESeq2)
library(edgeR)
library(bioDist)
library(biomaRt)
library(dplyr)
library(magrittr)
library(EnhancedVolcano)
library(ggplot2)
library(limma)
#library(R2HTML)
# set working environment to the location where current source file is saved into.
#setwd(dirname(rstudioapi::getSourceEditorContext()$path))
#include some functions adapted from ArrayAnalysis.org scripts
source("functions_ArrayAnalysis_v2.R")
WORK.DIR <- getwd()The following section will prepare the input data to be used in the analysis
#set wd one directory below to reach the input data
setwd('..')
WORK.DIR <- getwd()
#Obtain data from step 1
htxCount <- read.csv("1-data_preprocessing/output/htxCount.csv")
sampleLabels <- read.csv("1-data_preprocessing/output/sampleLabels.csv", header=FALSE)
# Set Working Directory back to current folder
#setwd(dirname(rstudioapi::getSourceEditorContext()$path))
setwd("2-differential_gene_expression_analysis")
WORK.DIR<- getwd()
#checking which samples have all zero values across all genes
#these sample should be removed otherwise there will be a problem when calculating estimate size factors
idx <- which(colSums(htxCount) == 0)
#CSMDRVXI MSM719ME are samples which has all zero values for all genes, so we remove them
htxCount <- htxCount[ , -idx]
#removing same samples from sample labels metadata
sampleLabels <- sampleLabels[-idx , ]
#Set column one as rownames
rownames(sampleLabels) <- sampleLabels[,1]
sampleLabels <- sampleLabels[,-1]
#add column names
colnames(sampleLabels) <- c( "sampleID", "biopsy_location","disease")
#check whether sample names are in same order
#all(colnames(htxCount) == rownames(sampleLabels2))
#select only biopsy_location and disease columns
sampleLabels<-sampleLabels[, c(2,3)]
sampleLabels$disease <- relevel(factor(sampleLabels$disease),ref="nonIBD")
#add an experimental group variable to sampleLabels
sampleLabels$group <- as.factor(paste(sampleLabels$disease,sampleLabels$biopsy_location,sep="_"))We will apply some filtering process to filter out genes in the input data
#remove genes which have all zero values across all samples then start DE analysis
nonzero <- rowSums(htxCount) > 0
htxCount %<>% .[nonzero,]
#############################CPM FILTERING#############################################
#aveLogCPM function computes average log2 counts-per-million for each row of counts.
#the below function is similar to log2(rowMeans(cpm(y, ...)))
mean_log_cpm = aveLogCPM(htxCount)
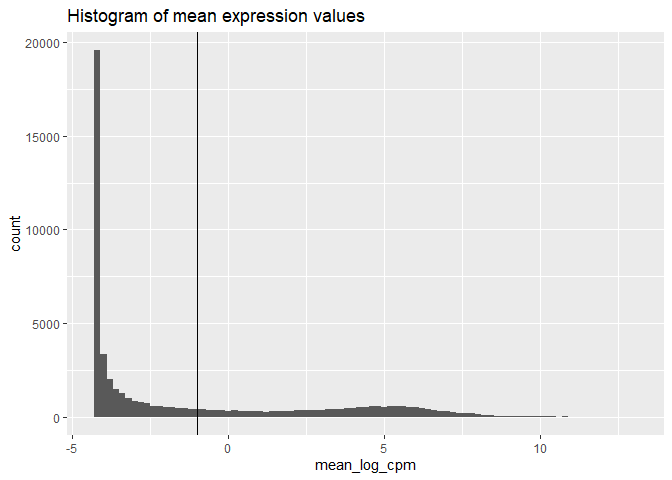
# We plot the distribution of average log2 CPM values to verify that our chosen presence threshold is appropriate. The distribution is expected to be bi modal, with a low-abundance peak representing non-expressed genes and a high-abundance peak representing expressed genes. The chosen threshold should separate the two peaks of the bi modal distribution.
filter_threshold <- -1# we can try different threshold values
#jpeg(file="avgLogCpmDist.jpeg")#if you want to save the histogram uncomment the following command
ggplot() + aes(x=mean_log_cpm) +
geom_histogram(binwidth=0.2) +
geom_vline(xintercept=filter_threshold) +
ggtitle("Histogram of mean expression values")#dev.off()#to save the plot to the file
#Having chosen our threshold, lets pick the subset of genes whose average expression passes that threshold.
keep_genes <- mean_log_cpm >= filter_threshold
htxCount <- htxCount[keep_genes,]
#dim(htxCount)#to check dimension of the data
###############################################################################In the following section differential gene expression analysis will be performed using DESeq2 package
# First create a DESeqDataSet object
#(non-intercept) statistical model based on the disease and biopsy_location, group column represent both of them
dds <- DESeqDataSetFromMatrix(countData = htxCount, colData=sampleLabels, design= ~0 + group)
#estimate the size factors
#To perform the median of ratios method of normalization, DESeq2 has a single estimateSizeFactors() function that will generate size factors for us.
dds <- estimateSizeFactors(dds)
#normalize the data (here for Quality Control(QC) plotting)
#QC plotting is optional
norm <- counts(dds,normalize=TRUE)
#create a 2logged data for original object (here for QC plotting)
datlog <- log(htxCount+1,2)
#create a 2logged norm object (here for QC plotting)
normlog <- log(norm+1,2)
#for QC remove genes that have not been measured in any sample in the experiment
datlogQC <- datlog[rowSums(datlog)!=0,]
normlogQC <- normlog[rowSums(normlog)!=0,]
#create QC plots for raw data, colored by different variables
factors <- c("disease","biopsy_location","group")
if(!dir.exists("QCraw")) dir.create("QCraw")
setwd(paste(WORK.DIR,"QCraw",sep="/"))
png("sizefactors.png")
plot(sizeFactors(dds),type='h',lwd=5,ylim=c(0,max(sizeFactors(dds))),col="darkblue")
dev.off()## png
## 2
createQCPlots(datlogQC, factors, Table=sampleLabels, normMeth="", postfix="")
setwd("..")
#create QC plots for normalized data colored by different variables
if(!dir.exists("QCnorm")) dir.create("QCnorm")
setwd(paste(WORK.DIR,"QCnorm",sep="/"))
createQCPlots(normlogQC, factors, Table=sampleLabels, normMeth="DESeq", postfix="")
setwd("..")
#sample MSM719M9 is an outlier remove it from dataset
#sample HSM5FZAZ is an outlier remove it from dataset
htxCount <- htxCount[,-match(c("MSM719M9","HSM5FZAZ"),colnames(htxCount))]
sampleLabels <- sampleLabels[-match(c("MSM719M9","HSM5FZAZ"),rownames(sampleLabels)),]
#doublecheck whether the order of the samples in sampleLabels and htxCount data still match
#sum(rownames(sampleLabels) == colnames(htxCount))==dim(sampleLabels)[1]
###REDO ALL STEPS AS ABOVE FOR FILTERED OUT DATASET
dds <- DESeqDataSetFromMatrix(countData = htxCount, colData=DataFrame(sampleLabels), design= ~0 + group)
dds <- estimateSizeFactors(dds)
norm <- counts(dds,normalize=TRUE)
datlog <- log(htxCount+1,2)
normlog <- log(norm+1,2)
datlogQC <- datlog[rowSums(datlog)!=0,]
normlogQC <- normlog[rowSums(normlog)!=0,]
#create QC plots for raw data, coloured by different variables
if(!dir.exists("QCraw2")) dir.create("QCraw2")
setwd(paste(WORK.DIR,"QCraw2",sep="/"))
png("sizefactors2.png")
plot(sizeFactors(dds),type='h',lwd=5,ylim=c(0,max(sizeFactors(dds))),col="darkblue")
dev.off()## png
## 2
createQCPlots(datlogQC, factors, Table=sampleLabels, normMeth="", postfix="")
setwd("..")
#create QC plots for normalized data coloured by different variables
if(!dir.exists("QCnorm2")) dir.create("QCnorm2")
setwd(paste(WORK.DIR,"QCnorm2",sep="/"))
createQCPlots(normlogQC, factors, Table=sampleLabels, normMeth="DESeq", postfix="")
setwd("..")
################################################################################
#######################statistical modelling####################################
################################################################################
#set directory for stat output
if(!dir.exists("statsmodel")) dir.create("statsmodel")
setwd(paste(WORK.DIR,"statsmodel",sep="/"))
#run differential analysis
dds <- DESeq(dds)
cont.matrix <- makeContrasts(
#CD disease on ileum and rectum
CD_Ileum_vs_nonIBD_Ileum = groupCD_Ileum - groupnonIBD_Ileum,
CD_Rectum_vs_nonIBD_Rectum = groupCD_Rectum - groupnonIBD_Rectum,
#UC disease on ileum and rectum
UC_Ileum_vs_nonIBD_Ileum = groupUC_Ileum - groupnonIBD_Ileum,
UC_Rectum_vs_nonIBD_Rectum = groupUC_Rectum - groupnonIBD_Rectum,
#UC and CD disease comparison
UC_Ileum_vs_CD_Illeum = groupUC_Ileum - groupCD_Ileum,
UC_Rectum_vs_CD_Rectum = groupUC_Rectum - groupCD_Rectum,
#biopsy location comparisons
CD_Rectum_vs_CD_Ileum = (groupCD_Rectum - groupnonIBD_Rectum) - (groupCD_Ileum - groupnonIBD_Ileum),
UC_Rectum_vs_UC_Ileum = (groupUC_Rectum - groupnonIBD_Rectum) - (groupUC_Ileum - groupnonIBD_Ileum),
levels = resultsNames(dds)
)
#extract resulting contrasts based on the model, and save those in a table; also save some graphical representations
#the function results() is called from within the saveStatOutputDESeq2 function to compute the contrasts
files <- saveStatOutputDESeq2(cont.matrix,dds,postfix="",annotation=NULL)## --[[ Saving table for coefficient CD_Ileum_vs_nonIBD_Ileum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient CD_Rectum_vs_nonIBD_Rectum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient UC_Ileum_vs_nonIBD_Ileum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient UC_Rectum_vs_nonIBD_Rectum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient UC_Ileum_vs_CD_Illeum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient UC_Rectum_vs_CD_Rectum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient CD_Rectum_vs_CD_Ileum ]]--
## ----[[ 0 probes with NA estimates removed ]]
## --[[ Saving table for coefficient UC_Rectum_vs_UC_Ileum ]]--
## ----[[ 0 probes with NA estimates removed ]]
#create summary table of the contrast results
createPvalTab(files,postfix="",namePVal="pvalue",nameAdjPVal="padj",nameFC="FoldChange",nameLogFC="log2FoldChange",html=TRUE)## --[[ Saving Summary_tables.tab ]]--
## --[[ Saving Summary_tables.html ]]--
setwd("..")Differential gene expression analysis results is visualized by volcano plots
#set the wd to the result folder statsmodel
setwd(paste(WORK.DIR,"statsmodel",sep="/"))
readFilePath <- paste(WORK.DIR,"statsmodel",sep="/")
# create an empty list
plot_list = list()
for(i in 1:length(files))
{
#read file
splitted <- strsplit(files[i], split = "_")[[1]]
title <- splitted [2:6]
title <- paste (splitted[2],splitted[3],splitted[4],splitted[5],splitted[6],sep=" ")
title <- (strsplit(title, split = "\\.")[[1]])[1]
tab <- read.delim(paste(readFilePath, files[i],sep="/"),header=TRUE,as.is=TRUE)
p<-EnhancedVolcano(tab , lab = tab$X, labSize = 3, title = title,
x = 'log2FoldChange', y = 'pvalue', pCutoff = 0.05, FCcutoff = 0.58)
plot_list[[i]] = p
}
#path of the output folder
outFolder <- paste(WORK.DIR,"volcano_plots",sep="/")
#create folder if doesnt exist
if(!dir.exists(outFolder)) dir.create(outFolder)
for(i in 1:length(files)) {
file_name = paste(outFolder,"/",files[i],".png",sep="")
png(file_name)
print(plot_list[[i]])
dev.off()
} ##Print session info and remove large datasets:
##Print session info:
sessionInfo()## R version 4.2.2 (2022-10-31 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 19044)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_Netherlands.utf8 LC_CTYPE=English_Netherlands.utf8
## [3] LC_MONETARY=English_Netherlands.utf8 LC_NUMERIC=C
## [5] LC_TIME=English_Netherlands.utf8
##
## attached base packages:
## [1] parallel stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] R2HTML_2.3.3 gdata_2.18.0.1
## [3] gplots_3.1.3 EnhancedVolcano_1.16.0
## [5] ggrepel_0.9.3 ggplot2_3.4.1
## [7] magrittr_2.0.3 dplyr_1.1.0
## [9] biomaRt_2.54.0 bioDist_1.70.0
## [11] KernSmooth_2.23-20 edgeR_3.40.2
## [13] limma_3.54.2 DESeq2_1.38.3
## [15] SummarizedExperiment_1.28.0 Biobase_2.58.0
## [17] MatrixGenerics_1.10.0 matrixStats_0.63.0
## [19] baySeq_2.32.0 abind_1.4-5
## [21] GenomicRanges_1.50.2 GenomeInfoDb_1.34.9
## [23] IRanges_2.32.0 S4Vectors_0.36.2
## [25] BiocGenerics_0.44.0 rstudioapi_0.14
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-7 bit64_4.0.5 filelock_1.0.2
## [4] RColorBrewer_1.1-3 progress_1.2.2 httr_1.4.5
## [7] tools_4.2.2 utf8_1.2.3 R6_2.5.1
## [10] DBI_1.1.3 colorspace_2.1-0 withr_2.5.0
## [13] tidyselect_1.2.0 prettyunits_1.1.1 bit_4.0.5
## [16] curl_5.0.0 compiler_4.2.2 cli_3.6.0
## [19] xml2_1.3.3 DelayedArray_0.24.0 labeling_0.4.2
## [22] caTools_1.18.2 scales_1.2.1 rappdirs_0.3.3
## [25] stringr_1.5.0 digest_0.6.31 rmarkdown_2.20
## [28] XVector_0.38.0 pkgconfig_2.0.3 htmltools_0.5.4
## [31] highr_0.10 dbplyr_2.3.1 fastmap_1.1.1
## [34] rlang_1.0.6 RSQLite_2.3.0 farver_2.1.1
## [37] generics_0.1.3 gtools_3.9.4 BiocParallel_1.32.5
## [40] RCurl_1.98-1.10 GenomeInfoDbData_1.2.9 Matrix_1.5-3
## [43] Rcpp_1.0.10 munsell_0.5.0 fansi_1.0.4
## [46] lifecycle_1.0.3 stringi_1.7.12 yaml_2.3.7
## [49] zlibbioc_1.44.0 BiocFileCache_2.6.1 grid_4.2.2
## [52] blob_1.2.3 crayon_1.5.2 lattice_0.20-45
## [55] Biostrings_2.66.0 annotate_1.76.0 hms_1.1.2
## [58] KEGGREST_1.38.0 locfit_1.5-9.7 knitr_1.42
## [61] pillar_1.8.1 geneplotter_1.76.0 codetools_0.2-19
## [64] XML_3.99-0.13 glue_1.6.2 evaluate_0.20
## [67] BiocManager_1.30.20 png_0.1-8 vctrs_0.5.2
## [70] gtable_0.3.1 cachem_1.0.7 xfun_0.37
## [73] xtable_1.8-4 tibble_3.1.8 AnnotationDbi_1.60.0
## [76] memoise_2.0.1 ellipsis_0.3.2
##Remove data objects which are not needed for further processing:
rm(list=setdiff(ls(), "tab"))#Jupyter Notebook file
if(!"devtools" %in% installed.packages()) BiocManager::install("devtools")
devtools::install_github("mkearney/rmd2jupyter", force=TRUE)##
## ── R CMD build ─────────────────────────────────────────────────────────────────
## checking for file 'C:\Users\duygu\AppData\Local\Temp\RtmpCO0e4d\remotes375c71eb78ad\mkearney-rmd2jupyter-d2bd2aa/DESCRIPTION' ... ✔ checking for file 'C:\Users\duygu\AppData\Local\Temp\RtmpCO0e4d\remotes375c71eb78ad\mkearney-rmd2jupyter-d2bd2aa/DESCRIPTION'
## ─ preparing 'rmd2jupyter':
## checking DESCRIPTION meta-information ... ✔ checking DESCRIPTION meta-information
## ─ checking for LF line-endings in source and make files and shell scripts
## ─ checking for empty or unneeded directories
## Omitted 'LazyData' from DESCRIPTION
## ─ building 'rmd2jupyter_0.1.0.tar.gz'
##
##
library(devtools)
library(rmd2jupyter)
#setwd(dirname(rstudioapi::getSourceEditorContext()$path))
rmd2jupyter("DEanalysis.Rmd")