citation("[package]")Setting time limits is an important consideration when tuning unreliable or unstable learning algorithms and when working on shared computing resources. The mlr3 ecosystem provides several mechanisms for setting time constraints for individual learners, tuning processes, and nested resampling.
+ +This section demonstrates how to impose time constraints using a support vector machine (SVM) as an illustrative example.
+library(mlr3verse)
+
+learner = lrn("classif.svm")Applying timeouts to the $train() and $predict() functions is essential for managing learners that may operate indefinitely. These time constraints are set independently for both the training and prediction stages. Generally, training a learner consumes more time than prediction. Certain learners, like k-nearest neighbors, lack a distinct training phase and require a timeout only during prediction. For the SVM’s training, we set a 10-second limit.
learner$timeout = c(train = 10, predict = Inf)To effectively terminate the process if necessary, it’s important to run the training and prediction within a separate R process. The callr package is recommended for this encapsulation, as it tends to be more reliable than the evaluate package, especially for terminating externally compiled code.
+learner$encapsulate = c(train = "callr", predict = "callr")Note that using callr increases the runtime due to the overhead of starting an R process. Additionally, it’s advisable to specify a fallback learner, such as "classif.featureless", to provide baseline predictions in case the primary learner is terminated.
learner$fallback = lrn("classif.featureless")These time constraints are now integrated into the training, resampling, and benchmarking processes. For more information on encapsulation and fallback learners, see the mlr3book. The next section will focus on setting time limits for the entire tuning process.
+When working with high-performance computing clusters, jobs are often bound by strict time constraints. Exceeding these limits results in the job being terminated and the loss of any results generated. Therefore, it’s important to ensure that the tuning process is designed to adhere to these time constraints.
+The trm("runtime") controls the duration of the tuning process. We must take into account that the terminator can only check if the time limit is reached between batches. We must therefore set the time lower than the runtime of the job. How much lower depends on the runtime or time limit of the individual learners. The last batch should be able to finish before the time limit of the cluster is reached.
terminator = trm("run_time", secs = 60)
+
+instance = ti(
+ task = tsk("sonar"),
+ learner = learner,
+ resampling = rsmp("cv", folds = 3),
+ measures = msr("classif.ce"),
+ terminator = terminator
+)With these settings, our tuning operation is configured to run for 60 seconds, while individual learners are set to terminate after 10 seconds. This approach ensures the tuning process is efficient and adheres to the constraints imposed by the high-performance computing cluster.
+When using nested resampling, time constraints become more complex as they are applied across various levels. As before, the time limit for an individual learner during the tuning is set with $timeout. The time limit for the tuning processes in the auto tuners is controlled with the trm("runtime"). It’s important to note that once the auto tuner enters the final phase of fitting the model and making predictions on the outer test set, the time limit governed by the terminator no longer applies. Additionally, the time limit previously set on the learner is temporarily deactivated, allowing the auto tuner to complete its task uninterrupted. However, a separate time limit can be assigned to each auto tuner using $timeout. This limit encompasses not only the tuning phase but also the time required for fitting the final model and predictions on the outer test set.
The best way to show this is with an example. We set the time limit for an individual learner to 10 seconds.
+learner$timeout = c(train = 10, predict = Inf)
+learner$encapsulate = c(train = "callr", predict = "callr")
+learner$fallback = lrn("classif.featureless")Next, we give each auto tuner 60 seconds to finish the tuning process.
+terminator = trm("run_time", secs = 60)Furthermore, we impose a 120-second limit for resampling each auto tuner. This effectively divides the time allocation, with around 60 seconds for tuning and another 60 seconds for final model fitting and predictions on the outer test set.
+at = auto_tuner(
+ tuner = tnr("random_search"),
+ learner = learner,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ terminator = trm("run_time", secs = 60)
+)
+
+at$timeout = c(train = 100, predict = 20)
+at$encapsulate = c(train = "callr", predict = "callr")
+at$fallback = lrn("classif.featureless")In total, the entire nested resampling process is designed to be completed within 10 minutes (120 seconds multiplied by 5 folds).
+rr = resample(task, at, rsmp("cv", folds = 5))We delved into the setting of time constraints across different levels in the mlr3 ecosystem. From individual learners to the complexities of nested resampling, we’ve seen how effectively managing time limits can significantly enhance the efficiency and reliability of machine learning workflows. By utilizing the trm("runtime") for tuning processes and setting $timeout for individual learners and auto tuners, we can ensure that our machine learning tasks are not only effective but also adhere to the practical time constraints of shared computational resources. For more information, see also the error handling section in the mlr3book.
In the realm of data science, machine learning frameworks play an important role in streamlining and accelerating the development of analytical workflows. Among these, tidymodels and mlr3 stand out as prominent tools within the R community. They provide a unified interface for data preprocessing, model training, resampling and tuning. The streamlined and accelerated development process, while efficient, typically results in a trade-off concerning runtime performance. This article undertakes a detailed comparison of the runtime efficiency of tidymodels and mlr3, focusing on their performance in training, resampling, and tuning machine learning models. Specifically, we assess the time efficiency of these frameworks in running the rpart::rpart() and ranger::ranger() models, using the Sonar dataset as a test case. Additionally, the study delves into analyzing the runtime overhead of these frameworks by comparing their performance against training the models without a framework. Through this comparative analysis, the article aims to provide valuable insights into the operational trade-offs of using these advanced machine learning frameworks in practical data science applications.
We employ the microbenchmark package to measure the time required for training, resampling, and tuning models. This benchmarking process is applied to the Sonar dataset using the rpart and ranger algorithms.
library("mlr3verse")
+library("tidymodels")
+library("microbenchmark")
+
+task = tsk("sonar")
+data = task$data()
+formula = Class ~ .To ensure the robustness of our results, each function call within the benchmark is executed 100 times in a randomized sequence. The microbenchmark package then provides us with detailed insights, including the median, lower quartile, and upper quartile of the runtimes. To further enhance the reliability of our findings, we execute the benchmark on a cluster. Each run of microbenchmark is repeated 100 times, with different seeds applied for each iteration. Resulting in a total of 10,000 function calls of each command. The computing environment for each worker in the cluster consists of 3 cores and 12 GB of RAM. For transparency and reproducibility, the examples of the code used for this experiment are provided as snippets in the article. The complete code, along with all details of the experiment, is available in our public repository, mlr-org/mlr-benchmark.
It’s important to note that our cluster setup is not specifically optimized for single-core performance. Consequently, executing the same benchmark on a local machine with might yield faster results.
+Our benchmark starts with the fundamental task of model training. To facilitate a direct comparison, we have structured our presentation into two distinct segments. On the left, we demonstrate the initialization of the rpart model, employing both mlr3 and tidymodels frameworks. The rpart model is a decision tree classifier, which is a simple and fast-fitting algorithm for classification tasks. Simultaneously, on the right, we turn our attention to the initialization of the ranger model, known for its efficient implementation of the random forest algorithm. Our aim is to mirror the configuration as closely as possible across both frameworks, maintaining consistency in parameters and settings.
# tidymodels
+tm_mod = decision_tree() %>%
+ set_engine("rpart",
+ xval = 0L) %>%
+ set_mode("classification")
+
+# mlr3
+learner = lrn("classif.rpart",
+ xval = 0L)
+# tidymodels
+tm_mod = rand_forest(trees = 1000L) %>%
+ set_engine("ranger",
+ num.threads = 1L,
+ seed = 1) %>%
+ set_mode("classification")
+
+# mlr3
+learner = lrn("classif.ranger",
+ num.trees = 1000L,
+ num.threads = 1L,
+ seed = 1,
+ verbose = FALSE,
+ predict_type = "prob")We measure the runtime for the train functions within each framework. The result of the train function is a trained model in both frameworks. In addition, we invoke the rpart() and ranger() functions to establish a baseline for the minimum achievable runtime. This allows us to not only assess the efficiency of the train functions in each framework but also to understand how they perform relative to the base packages.
# tidymodels train
+fit(tm_mod, formula, data = data)
+
+# mlr3 train
+learner$train(task)When training an rpart model, tidymodels demonstrates superior speed, outperforming mlr3 (Table 1). Notably, the mlr3 package requires approximately twice the time compared to the baseline.
A key observation from our results is the significant relative overhead when using a framework for rpart model training. Given that rpart inherently requires a shorter training time, the additional processing time introduced by the frameworks becomes more pronounced. This aspect highlights the trade-off between the convenience offered by these frameworks and their impact on runtime for quicker tasks.
Conversely, when we shift our focus to training a ranger model, the scenario changes (Table 2). Here, the runtime performance of ranger is strikingly similar across both tidymodels and mlr3. This equality in execution time can be attributed to the inherently longer training duration required by ranger models. As a result, the relative overhead introduced by either framework becomes minimal, effectively diminishing in the face of the more time-intensive training process. This pattern suggests that for more complex or time-consuming tasks, the choice of framework may have a less significant impact on overall runtime performance.
rpart depending on the framework.
+| Framework | +LQ | +Median | +UQ | +
|---|---|---|---|
| base | +11 | +11 | +12 | +
| mlr3 | +23 | +23 | +24 | +
| tidymodels | +18 | +18 | +19 | +
ranger depending on the framework.
+| Framework | +LQ | +Median | +UQ | +
|---|---|---|---|
| base | +286 | +322 | +347 | +
| mlr3 | +301 | +335 | +357 | +
| tidymodels | +310 | +342 | +362 | +
We proceed to evaluate the runtime performance of the resampling functions within both frameworks, specifically under conditions without parallelization. This step involves the generation of resampling splits, including 3-fold, 6-fold, and 9-fold cross-validation. Additionally, we run a 100 times repeated 3-fold cross-validation.
+We generate the same resampling splits for both frameworks. This consistency is key to ensuring that any observed differences in runtime are attributable to the frameworks themselves, rather than variations in the resampling process.
+In our pursuit of a fair and balanced comparison, we address certain inherent differences between the two frameworks. Notably, tidymodels inherently includes scoring of the resampling results as part of its process. To align the comparison, we replicate this scoring step in mlr3, thus maintaining a level field for evaluation. Furthermore, mlr3 inherently saves predictions during the resampling process. To match this, we activate the saving of the predictions in tidymodels.
# tidymodels resample
+control = control_grid(save_pred = TRUE)
+metrics = metric_set(accuracy)
+
+tm_wf =
+ workflow() %>%
+ add_model(tm_mod) %>%
+ add_formula(formula)
+
+fit_resamples(tm_wf, folds, metrics = metrics, control = control)
+
+# mlr3 resample
+measure = msr("classif.acc")
+
+rr = resample(task, learner, resampling)
+rr$score(measure)When resampling the fast-fitting rpart model, mlr3 demonstrates a notable edge in speed, as detailed in Table 3. In contrast, when it comes to resampling the more computationally intensive ranger models, the performance of tidymodels and mlr3 converges closely (Table 4). This parity in performance is particularly noteworthy, considering the differing internal mechanisms and optimizations of tidymodels and mlr3. A consistent trend observed across both frameworks is a linear increase in runtime proportional to the number of folds in cross-validation (Figure 1).
rpart depending on the framework and resampling strategy.
+| Framework | +Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|---|
| mlr3 | +cv3 | +188 | +196 | +210 | +
| tidymodels | +cv3 | +233 | +242 | +257 | +
| mlr3 | +cv6 | +343 | +357 | +379 | +
| tidymodels | +cv6 | +401 | +415 | +436 | +
| mlr3 | +cv9 | +500 | +520 | +548 | +
| tidymodels | +cv9 | +568 | +588 | +616 | +
| mlr3 | +rcv100 | +15526 | +16023 | +16777 | +
| tidymodels | +rcv100 | +16409 | +16876 | +17527 | +
ranger depending on the framework and resampling strategy.
+| Framework | +Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|---|
| mlr3 | +cv3 | +923 | +1004 | +1062 | +
| tidymodels | +cv3 | +916 | +981 | +1023 | +
| mlr3 | +cv6 | +1990 | +2159 | +2272 | +
| tidymodels | +cv6 | +2089 | +2176 | +2239 | +
| mlr3 | +cv9 | +3074 | +3279 | +3441 | +
| tidymodels | +cv9 | +3260 | +3373 | +3453 | +
| mlr3 | +rcv100 | +85909 | +88642 | +91381 | +
| tidymodels | +rcv100 | +87828 | +88822 | +89843 | +
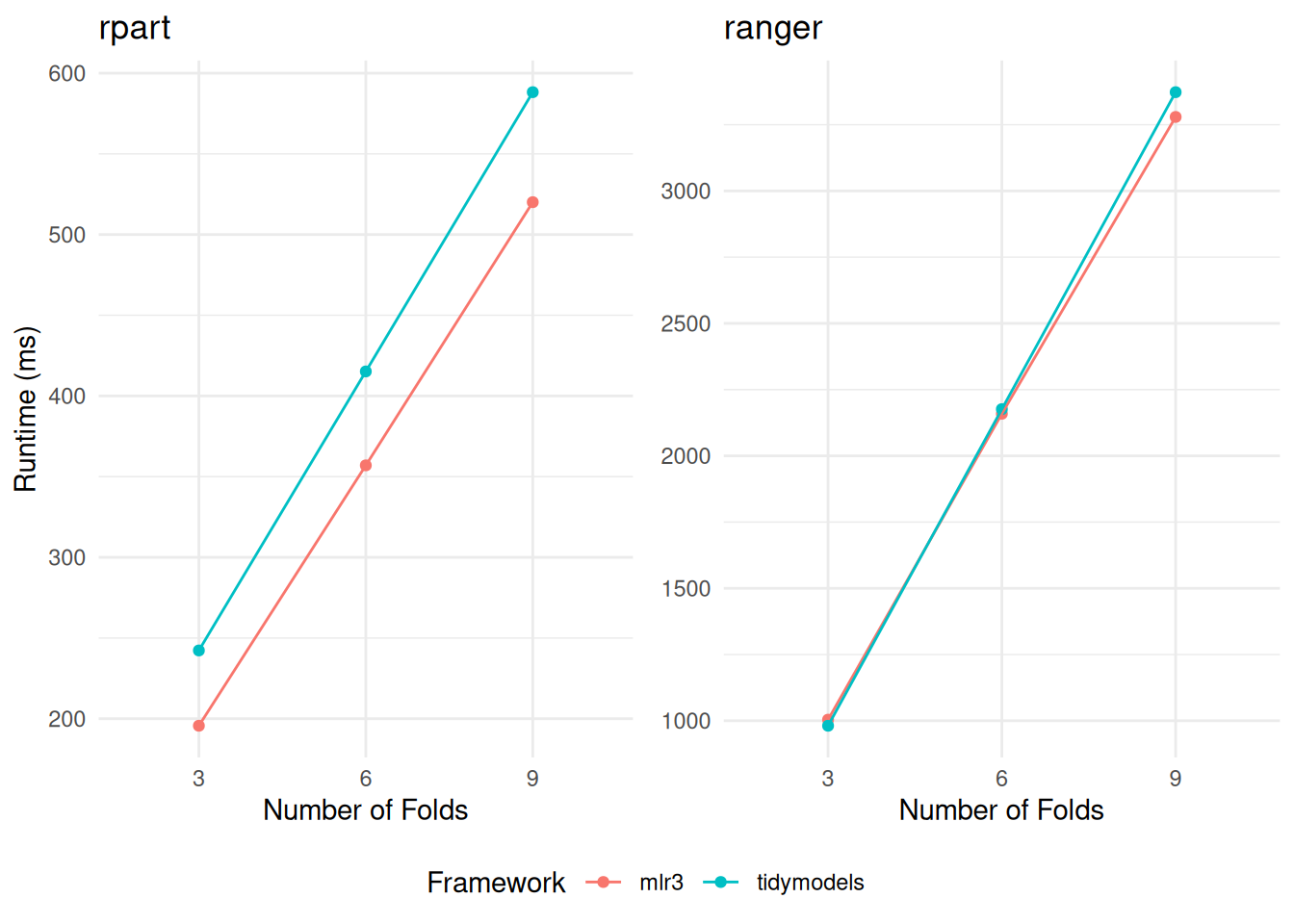 +
+rpart (displayed on the left) and ranger (on the right). The comparison encompasses variations across different frameworks and the number of folds in the cross-validation.
+We conducted a second set of resampling function tests, this time incorporating parallelization to explore its impact on runtime efficiency. In this phase, we utilized doFuture and doParallel as the primary parallelization packages for tidymodels, recognizing their robust support and compatibility. Meanwhile, for mlr3, the future package was employed to facilitate parallel processing.
Our findings, as presented in the respective tables (Table 5 and Table 6), reveal interesting dynamics about parallelization within the frameworks. When the number of folds in the resampling process is doubled, we observe only a marginal increase in the average runtime. This pattern suggests a significant overhead associated with initializing the parallel workers, a factor that becomes particularly influential in the overall efficiency of the parallelization process.
+In the case of the rpart model, the parallelization overhead appears to outweigh the potential speedup benefits, as illustrated in the left section of Figure 2. This result indicates that for less complex models like rpart, where individual training times are relatively short, the initialization cost of parallel workers may not be sufficiently offset by the reduced processing time per fold.
Conversely, for the ranger model, the utilization of parallelization demonstrates a clear advantage over the sequential version, as evidenced in the right section of Figure 2. This finding underscores that for more computationally intensive models like ranger, which have longer individual training times, the benefits of parallel processing significantly overcome the initial overhead of worker setup. This differentiation highlights the importance of considering the complexity and inherent processing time of models when deciding to implement parallelization strategies in these frameworks.
mlr3 with future and rpart depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +625 | +655 | +703 | +
| cv6 | +738 | +771 | +817 | +
| cv9 | +831 | +875 | +923 | +
| rcv100 | +8620 | +9043 | +9532 | +
mlr3 with future and ranger depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +836 | +884 | +943 | +
| cv6 | +1200 | +1249 | +1314 | +
| cv9 | +1577 | +1634 | +1706 | +
| rcv100 | +32047 | +32483 | +33022 | +
When paired with doFuture, tidymodels exhibits significantly slower runtime compared to the mlr3 package utilizing future (Table 7 and Table 8). We observed that tidymodels exports more data to the parallel workers, which notably exceeds that of mlr3. This substantial difference in data export could plausibly account for the observed slower runtime when using tidymodels on small tasks.
tidymodels with doFuture and rpart depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +2778 | +2817 | +3019 | +
| cv6 | +2808 | +2856 | +3033 | +
| cv9 | +2935 | +2975 | +3170 | +
| rcv100 | +9154 | +9302 | +9489 | +
tidymodels with doFuture and ranger depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +2982 | +3046 | +3234 | +
| cv6 | +3282 | +3366 | +3543 | +
| cv9 | +3568 | +3695 | +3869 | +
| rcv100 | +27546 | +27843 | +28166 | +
The utilization of the doParallel package demonstrates a notable improvement in handling smaller resampling tasks. In these scenarios, the resampling process consistently outperforms the mlr3 framework in terms of speed. However, it’s important to note that even with this enhanced performance, the doParallel package does not always surpass the efficiency of the sequential version, especially when working with the rpart model. This specific observation is illustrated in the left section of Figure 2.
tidymodels with doParallel and rpart depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +557 | +649 | +863 | +
| cv6 | +602 | +714 | +910 | +
| cv9 | +661 | +772 | +968 | +
| rcv100 | +10609 | +10820 | +11071 | +
tidymodels with doParallel and ranger depending on the resampling strategy.
+| Resampling | +LQ | +Median | +UQ | +
|---|---|---|---|
| cv3 | +684 | +756 | +948 | +
| cv6 | +1007 | +1099 | +1272 | +
| cv9 | +1360 | +1461 | +1625 | +
| rcv100 | +31205 | +31486 | +31793 | +
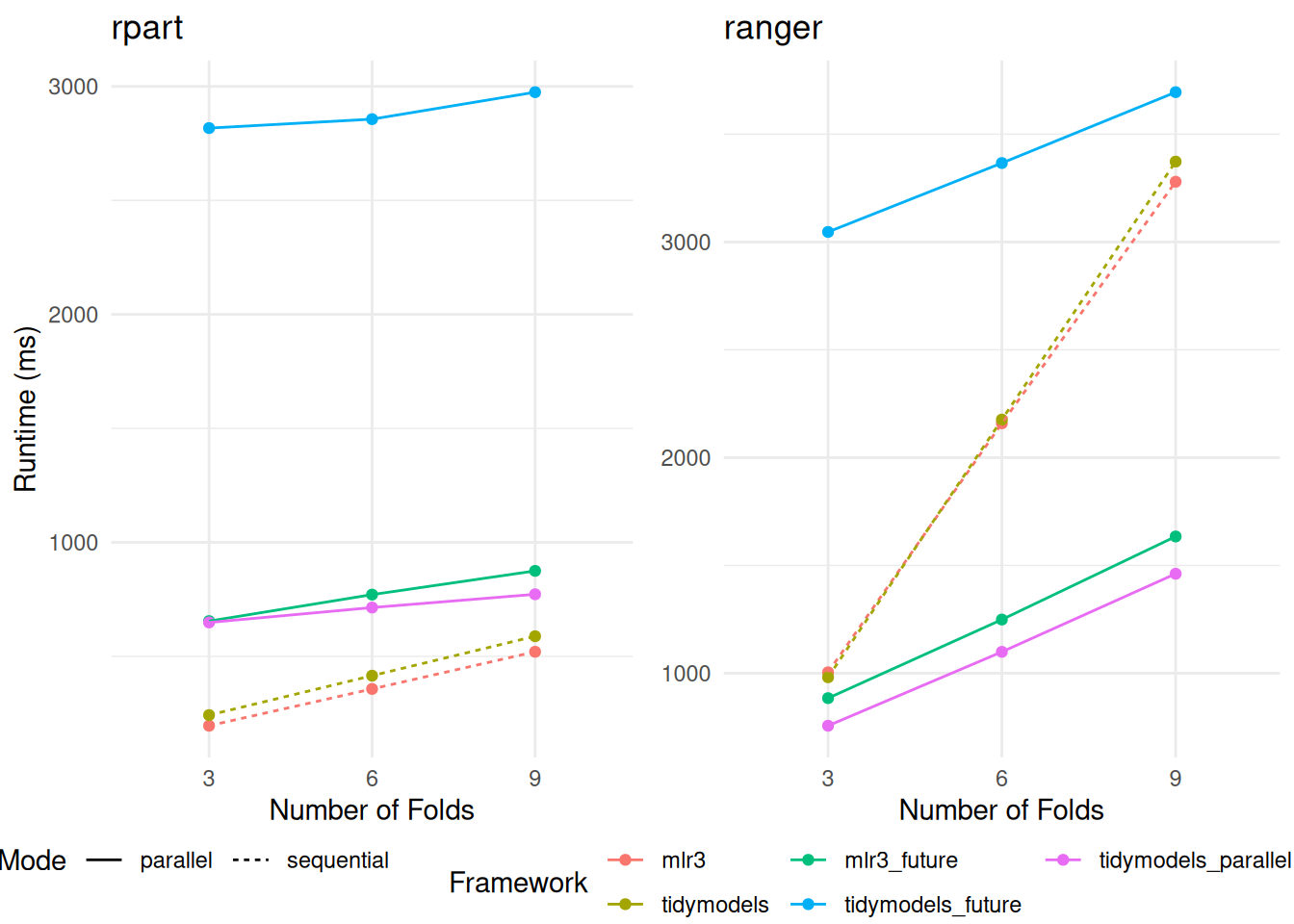 +
+rpart (displayed on the left) and ranger (on the right). The comparison encompasses variations across different frameworks, the number of folds in the cross-validation, and the implementation of parallelization.
+In the context of repeated cross-validation, our findings underscore the efficacy of parallelization (Figure 3). Across all frameworks tested, the adoption of parallel processing techniques yields a significant increase in speed. This enhancement is particularly noticeable in larger resampling tasks, where the demands on computational resources are more substantial.
+Interestingly, within these more extensive resampling scenarios, the doFuture package emerges as a more efficient option compared to doParallel. This distinction is important, as it highlights the relative strengths of different parallelization packages under varying workload conditions. While doParallel shows proficiency in smaller tasks, doFuture demonstrates its capability to handle larger, more complex resampling processes with greater speed and efficiency.
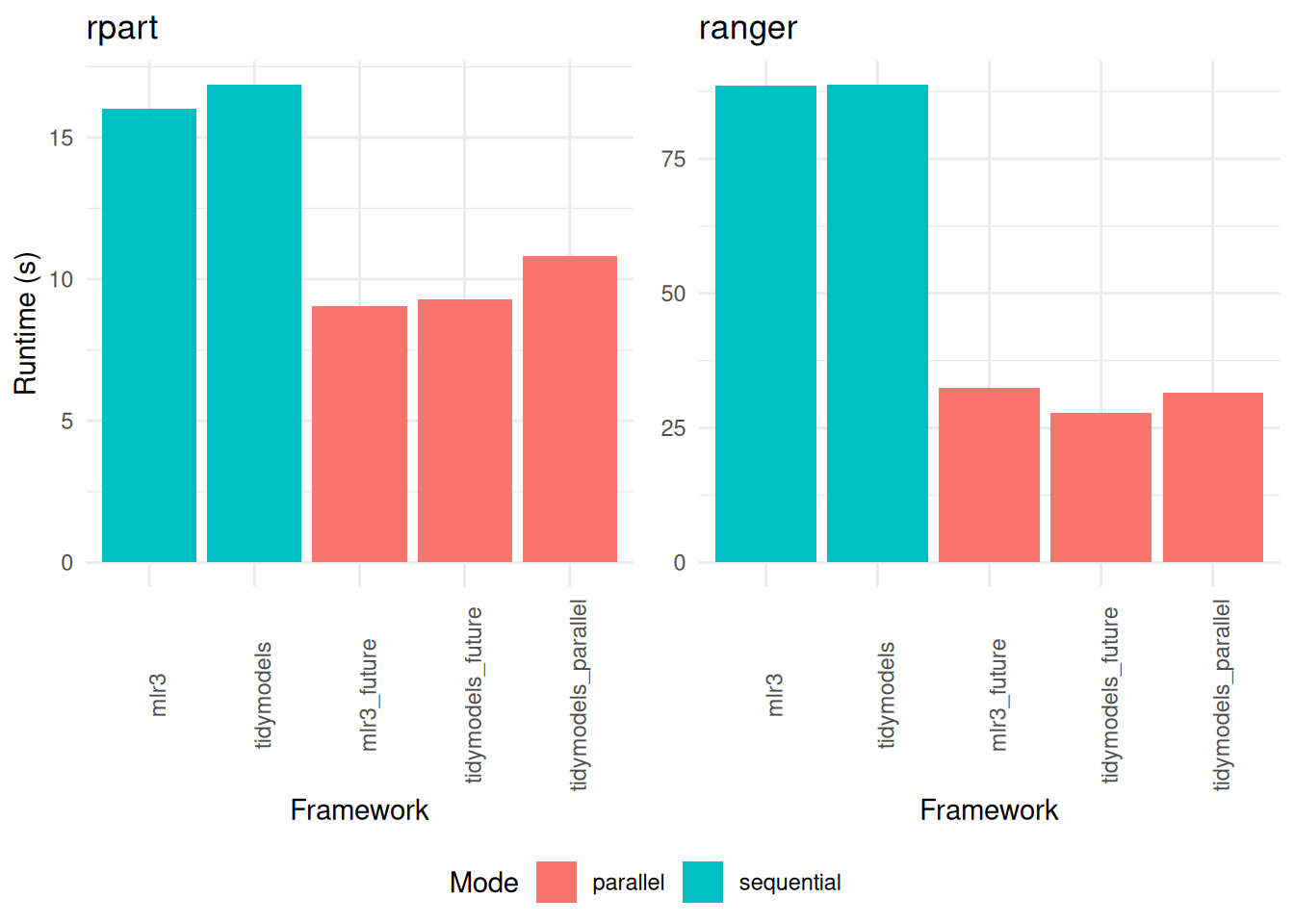 +
+rpart (displayed on the left) and ranger (on the right). The comparison encompasses variations across different frameworks and the implementation of parallelization.
+We then shift our focus to assessing the runtime performance of the tuning functions. In this phase, the tidymodels package is utilized to evaluate a predefined grid, comprising a specific set of hyperparameter configurations. To ensure a balanced and comparable analysis, we employ the "design_points" tuner from the mlr3tuning package. This approach allows us to evaluate the same grid within the mlr3 framework, maintaining consistency across both platforms. The grid used for this comparison contains 200 hyperparameter configurations each, for both the rpart and ranger models. This approach helps us to understand how each framework handles the optimization of model hyperparameters, a key aspect of building effective and efficient machine learning models.
# tidymodels
+tm_mod = decision_tree(
+ cost_complexity = tune()) %>%
+ set_engine("rpart",
+ xval = 0) %>%
+ set_mode("classification")
+
+tm_design = data.table(
+ cost_complexity = seq(0.1, 0.2, length.out = 200))
+
+# mlr3
+learner = lrn("classif.rpart",
+ xval = 0,
+ cp = to_tune())
+
+mlr3_design = data.table(
+ cp = seq(0.1, 0.2, length.out = 200))
+# tidymodels
+tm_mod = rand_forest(
+ trees = tune()) %>%
+ set_engine("ranger",
+ num.threads = 1L,
+ seed = 1) %>%
+ set_mode("classification")
+
+tm_design = data.table(
+ trees = seq(1000, 1199))
+
+# mlr3
+learner = lrn("classif.ranger",
+ num.trees = to_tune(1, 10000),
+ num.threads = 1L,
+ seed = 1,
+ verbose = FALSE,
+ predict_type = "prob")
+
+mlr3_design = data.table(
+ num.trees = seq(1000, 1199))We measure the runtime of the tune functions within each framework. Both the tidymodels and mlr3 frameworks are tasked with identifying the optimal hyperparameter configuration.
# tidymodels tune
+tune::tune_grid(
+ tm_wf,
+ resamples = resamples,
+ grid = design,
+ metrics = metrics)
+
+# mlr3 tune
+tuner = tnr("design_points", design = design, batch_size = nrow(design))
+mlr3tuning::tune(
+ tuner = tuner,
+ task = task,
+ learner = learner,
+ resampling = resampling,
+ measures = measure,
+ store_benchmark_result = FALSE)In our sequential tuning tests, mlr3 demonstrates a notable advantage in terms of speed. This finding is clearly evidenced in our results, as shown in Table Table 11 for the rpart model and Table Table 12 for the ranger model. The faster performance of mlr3 in these sequential runs highlights its efficiency in handling the tuning process without parallelization.
rpart depending on the framework.
+| Framework | +LQ | +Median | +UQ | +
|---|---|---|---|
| mlr3 | +27 | +27 | +28 | +
| tidymodels | +37 | +37 | +39 | +
ranger depending on the framework.
+| Framework | +LQ | +Median | +UQ | +
|---|---|---|---|
| mlr3 | +167 | +171 | +175 | +
| tidymodels | +194 | +195 | +196 | +
Concluding our analysis, we proceed to evaluate the runtime performance of the tune functions, this time implementing parallelization to enhance efficiency. For these runs, parallelization is executed on 3 cores.
+In the case of mlr3, we opt for the largest possible chunk size. This strategic choice means that all points within the tuning grid are sent to the workers in a single batch, effectively minimizing the overhead typically associated with parallelization. This approach is crucial in reducing the time spent in distributing tasks across multiple cores, thereby streamlining the tuning process. On the other hand, the tidymodels package also operates with the same chunk size, but this setting is determined and managed internally within the framework.
By conducting these parallelization tests, we aim to provide a deeper understanding of how each framework handles the distribution and management of computational tasks during the tuning process, particularly in a parallel computing environment. This final set of measurements is important in painting a complete picture of the runtime performance of the tune functions across both tidymodels and mlr3 under different operational settings.
options("mlr3.exec_chunk_size" = 200)Our analysis of the parallelized tuning functions reveals that the runtimes for mlr3 and tidymodels are remarkably similar. However, subtle differences emerge upon closer inspection. For instance, the mlr3 package exhibits a slightly faster performance when tuning the rpart model, as indicated in Table 13. In contrast, it falls marginally behind tidymodels in tuning the ranger model, as shown in Table 14.
Interestingly, when considering the specific context of a 3-fold cross-validation, the doParallel package outperforms doFuture in terms of speed, as demonstrated in Figure 4. This outcome suggests that the choice of parallelization package can have a significant impact on tuning efficiency, particularly in scenarios with a smaller number of folds.
A key takeaway from our study is the clear benefit of enabling parallelization, regardless of the chosen framework-backend combination. Activating parallelization consistently enhances performance, making it a highly recommended strategy for tuning machine learning models, especially in tasks involving extensive hyperparameter exploration or larger datasets. This conclusion underscores the value of parallel processing in modern machine learning workflows, offering a practical solution for accelerating model tuning across various computational settings.
+rpart depending on the framework.
+| Framework | +Backend | +LQ | +Median | +UQ | +
|---|---|---|---|---|
| mlr3 | +future | +11 | +12 | +12 | +
| tidymodels | +doFuture | +17 | +17 | +17 | +
| tidymodels | +doParallel | +13 | +13 | +13 | +
ranger depending on the framework.
+| Framework | +Backend | +LQ | +Median | +UQ | +
|---|---|---|---|---|
| mlr3 | +future | +54 | +55 | +55 | +
| tidymodels | +doFuture | +58 | +58 | +59 | +
| tidymodels | +doParallel | +54 | +54 | +55 | +
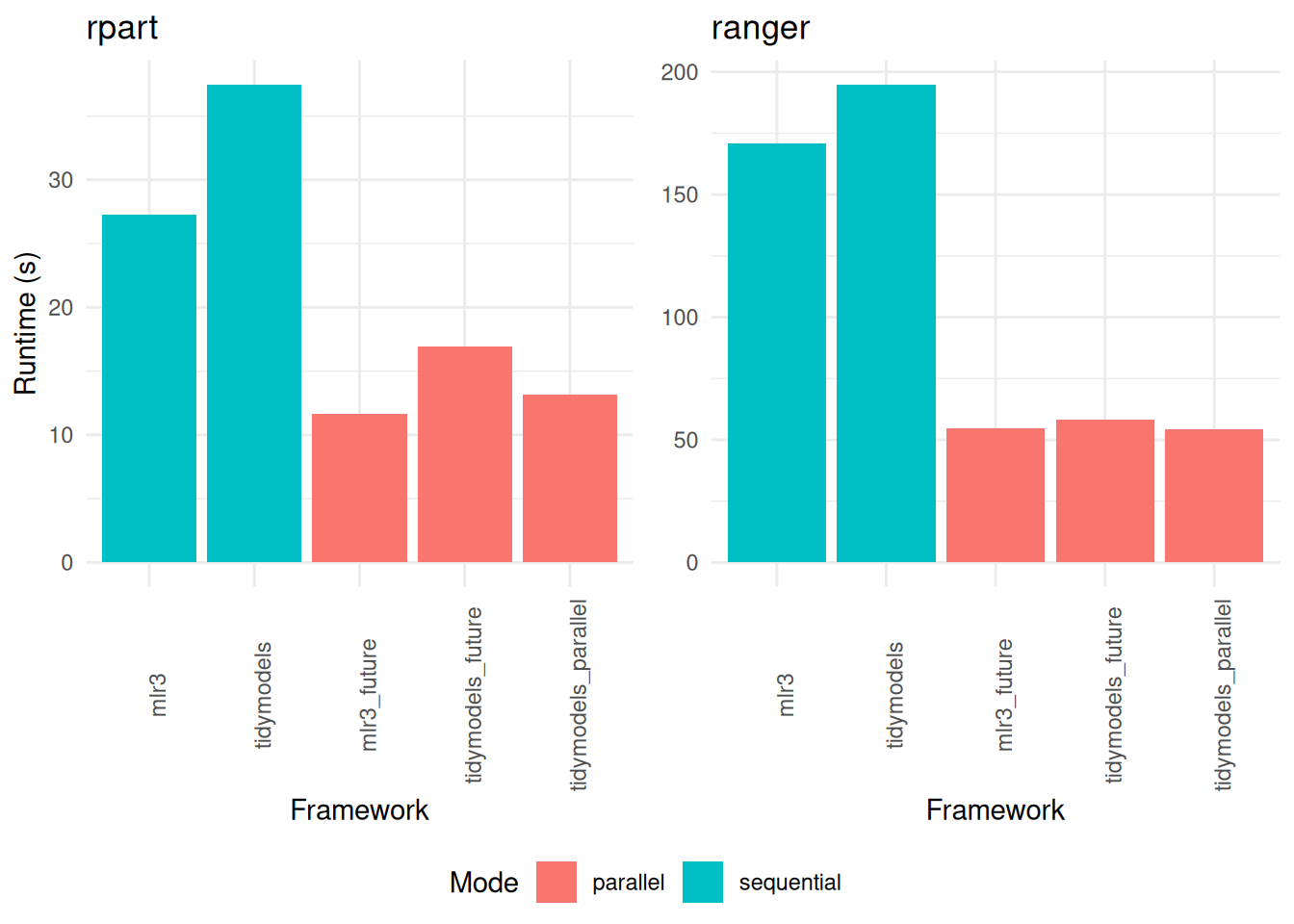 +
+rpart (displayed on the left) and ranger (on the right). The comparison encompasses variations across different frameworks and the implementation of parallelization.
+Our analysis reveals that both tidymodels and mlr3 exhibit comparable runtimes across key processes such as training, resampling, and tuning, each displaying its own set of strengths and efficiencies.
A notable observation is the relative overhead associated with using either framework, particularly when working with fast-fitting models like rpart. In these cases, the additional processing time introduced by the frameworks is more pronounced due to the inherently short training time of rpart models. This results in a higher relative overhead, reflecting the trade-offs between the convenience of a comprehensive framework and the directness of more basic approaches.
Conversely, when dealing with slower-fitting models such as ranger, the scenario shifts. For these more time-intensive models, the relative overhead introduced by the frameworks diminishes significantly. In such instances, the extended training times of the models absorb much of the frameworks’ inherent overhead, rendering it relatively negligible.
In summary, while there is no outright winner in terms of overall performance, the decision to use tidymodels or mlr3 should be informed by the specific requirements of the task at hand.
Here are some interesting reads regarding BART:
+BART R package tutorial (R. Sparapani, Spanbauer, and McCulloch 2021)We incorporated the survival BART model in mlr3extralearners and in this tutorial we will demonstrate how we can use packages like mlr3, mlr3proba and distr6 to more easily manipulate the output predictions to assess model convergence, validate our model (via several survival metrics), as well as perform model interpretation via PDPs (Partial Dependence Plots).
library(mlr3extralearners)
+library(mlr3pipelines)
+library(mlr3proba)
+library(distr6)
+library(BART) # 2.9.4
+library(dplyr)
+library(tidyr)
+library(tibble)
+library(ggplot2)We will use the Lung Cancer Dataset. We convert the time variable from days to months to ease the computational burden:
task_lung = tsk('lung')
+
+d = task_lung$data()
+# in case we want to select specific columns to keep
+# d = d[ ,colnames(d) %in% c("time", "status", "age", "sex", "ph.karno"), with = FALSE]
+d$time = ceiling(d$time/30.44)
+task_lung = as_task_surv(d, time = 'time', event = 'status', id = 'lung')
+task_lung$label = "Lung Cancer"BART implementation supports categorical features (factors). This results in different importance scores per each dummy level which doesn’t work well with mlr3. So features of type factor or character are not allowed and we leave it to the user to encode them as they please.BART implementation supports features with missing values. This is totally fine with mlr3 as well! In this example, we impute the features to show good ML practice.In our lung dataset, we encode the sex feature and perform model-based imputation with the rpart regression learner:
po_encode = po('encode', method = 'treatment')
+po_impute = po('imputelearner', lrn('regr.rpart'))
+pre = po_encode %>>% po_impute
+task = pre$train(task_lung)[[1]]
+task<TaskSurv:lung> (228 x 10): Lung Cancer
+* Target: time, status
+* Properties: -
+* Features (8):
+ - int (7): age, inst, meal.cal, pat.karno, ph.ecog, ph.karno, wt.loss
+ - dbl (1): sexNo missing values in our data:
+task$missings() time status age sex inst meal.cal pat.karno ph.ecog ph.karno wt.loss
+ 0 0 0 0 0 0 0 0 0 0 We partition the data to train and test sets:
+set.seed(42)
+part = partition(task, ratio = 0.9)We train the BART model and predict on the test set:
# default `ndpost` value: 1000. We reduce it to 50 to speed up calculations in this tutorial
+learner = lrn("surv.bart", nskip = 250, ndpost = 50, keepevery = 10, mc.cores = 10)
+learner$train(task, row_ids = part$train)
+p = learner$predict(task, row_ids = part$test)
+p<PredictionSurv> for 23 observations:
+ row_ids time status crank distr
+ 9 8 TRUE 66.19326 <list[1]>
+ 10 6 TRUE 98.43005 <list[1]>
+ 21 10 TRUE 54.82313 <list[1]>
+---
+ 160 13 FALSE 37.82089 <list[1]>
+ 163 10 FALSE 69.63534 <list[1]>
+ 194 8 FALSE 81.13678 <list[1]>See more details about BART’s parameters on the online documentation.
What kind of object is the predicted distr?
p$distrArrdist(23x31x50) Actually the $distr is an active R6 field - this means that some computation is required to create it. What the prediction object actually stores internally is a 3d survival array (can be used directly with no performance overhead):
dim(p$data$distr)[1] 23 31 50This is a more easy-to-understand and manipulate form of the full posterior survival matrix prediction from the BART package ((R. Sparapani, Spanbauer, and McCulloch 2021), pages 34-35).
Though we have optimized with C++ code the way the Arrdist object is constructed, calling the $distr field can be computationally taxing if the product of the sizes of the 3 dimensions above exceeds ~1 million. In our case, so the conversion to an
Arrdist via $distr will certainly not create performance issues.
An example using the internal prediction data: get all the posterior probabilities of the 3rd patient in the test set, at 12 months (1 year):
+p$data$distr[3, 12, ] [1] 0.26546909 0.27505937 0.21151435 0.46700513 0.26178380 0.24040003 0.29946469 0.52357780 0.40833108 0.40367780
+[11] 0.27027392 0.31781286 0.54151844 0.34460027 0.41826554 0.41866367 0.33694401 0.34511270 0.47244492 0.49423660
+[21] 0.42069678 0.20095489 0.48696980 0.48409357 0.35649439 0.47969355 0.16355660 0.33728105 0.40245228 0.42418033
+[31] 0.36336145 0.48181667 0.51858238 0.49635078 0.37238179 0.26694030 0.52219952 0.48992897 0.08572207 0.30306005
+[41] 0.33881682 0.33463870 0.29102074 0.43176131 0.38554545 0.38053756 0.36808776 0.13772665 0.21898264 0.14552514Working with the $distr interface and Arrdist objects is very efficient as we will see later for predicting survival estimates.
In survival analysis, , where
the survival function and
the cumulative distribution function (cdf). The latter can be interpreted as
risk or probability of death up to time .
We can verify the above from the prediction object:
+surv_array = 1 - distr6::gprm(p$distr, "cdf") # 3d array
+testthat::expect_equal(p$data$distr, surv_array)crank is the expected mortality (Sonabend, Bender, and Vollmer 2022) which is the sum of the predicted cumulative hazard function (as is done in random survival forest models). Higher values denote larger risk. To calculate crank, we need a survival matrix. So we have to choose which 3rd dimension we should use from the predicted survival array. This is what the which.curve parameter of the learner does:
learner$param_set$get_values()$which.curve[1] 0.5The default value ( quantile) is the median survival probability. It could be any other quantile (e.g.
). Other possible values for
which.curve are mean or a number denoting the exact posterior draw to extract (e.g. the last one, which.curve = 50).
Default score is the observed count of each feature in the trees (so the higher the score, the more important the feature):
+learner$param_set$values$importance[1] "count"learner$importance() sex meal.cal inst pat.karno ph.karno wt.loss age ph.ecog
+ 7.84 7.46 7.08 6.76 6.60 6.46 5.48 5.42 BART uses internally MCMC (Markov Chain Monte Carlo) to sample from the posterior survival distribution. We need to check that MCMC has converged, meaning that the chains have reached a stationary distribution that approximates the true posterior survival distribution (otherwise the predictions may be inaccurate, misleading and unreliable).
We use Geweke’s convergence diagnostic test as it is implemented in the BART R package. We choose 10 random patients from the train set to evaluate the MCMC convergence.
# predictions on the train set
+p_train = learner$predict(task, row_ids = part$train)
+
+# choose 10 patients from the train set randomly and make a list
+ids = as.list(sample(length(part$train), 10))
+
+z_list = lapply(ids, function(id) {
+ # matrix with columns => time points and rows => posterior draws
+ post_surv = 1 - t(distr6::gprm(p_train$distr[id], "cdf")[1,,])
+ BART::gewekediag(post_surv)$z # get the z-scores
+})
+
+# plot the z scores vs time for all patients
+dplyr::bind_rows(z_list) %>%
+ tidyr::pivot_longer(cols = everything()) %>%
+ mutate(name = as.numeric(name)) %>%
+ ggplot(aes(x = name, y = value)) +
+ geom_point() +
+ labs(x = "Time (months)", y = "Z-scores") +
+ # add critical values for a = 0.05
+ geom_hline(yintercept = 1.96, linetype = 'dashed', color = "red") +
+ geom_hline(yintercept = -1.96, linetype = 'dashed', color = "red") +
+ theme_bw(base_size = 14)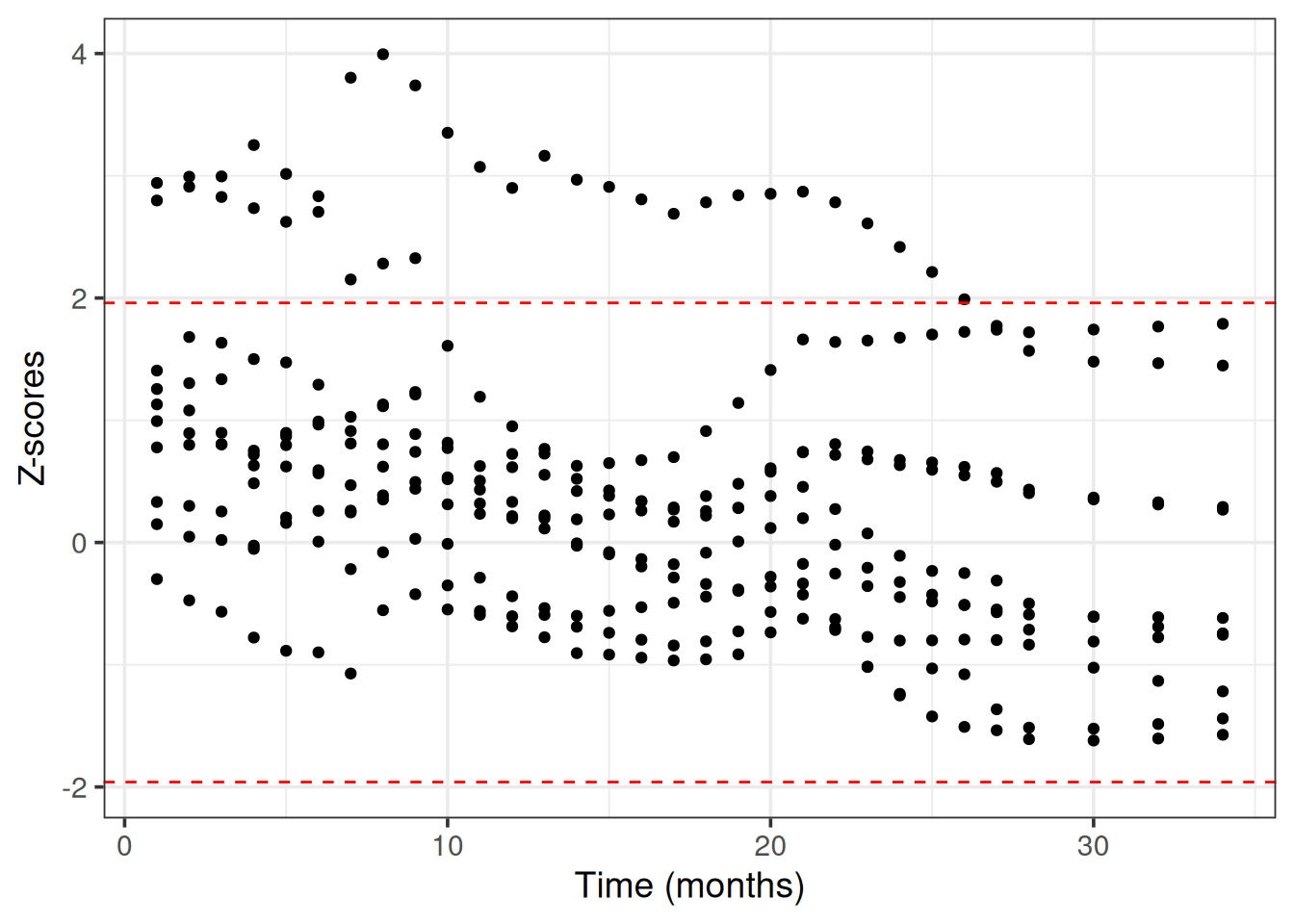
We will use the following survival metrics:
+distr)crank)For the first measure we will use the ERV (Explained Residual Variation) version, which standardizes the score against a Kaplan-Meier (KM) baseline (Sonabend et al. 2022). This means that values close to represent performance similar to a KM model, negative values denote worse performance than KM and
is the absolute best possible score.
measures = list(
+ msr("surv.graf", ERV = TRUE),
+ msr("surv.cindex", weight_meth = "G2", id = "surv.cindex.uno")
+)
+
+for (measure in measures) {
+ print(p$score(measure, task = task, train_set = part$train))
+} surv.graf
+-0.09950096
+surv.cindex.uno
+ 0.551951 All metrics use by default the median survival distribution from the 3d array, no matter what is the which.curve argument during the learner’s construction.
Performing resampling with the BART learner is very easy using mlr3.
We first stratify the data by status, so that in each resampling the proportion of censored vs un-censored patients remains the same:
task$col_roles$stratum = 'status'
+task$strata N row_id
+1: 165 1,2,4,5,7,8,...
+2: 63 3, 6,38,68,71,83,...rr = resample(task, learner, resampling = rsmp("cv", folds = 5), store_backends = TRUE)INFO [17:07:04.946] [mlr3] Applying learner 'surv.bart' on task 'lung' (iter 1/5)
+INFO [17:07:07.647] [mlr3] Applying learner 'surv.bart' on task 'lung' (iter 2/5)
+INFO [17:07:10.708] [mlr3] Applying learner 'surv.bart' on task 'lung' (iter 3/5)
+INFO [17:07:13.382] [mlr3] Applying learner 'surv.bart' on task 'lung' (iter 4/5)
+INFO [17:07:16.361] [mlr3] Applying learner 'surv.bart' on task 'lung' (iter 5/5)No errors or warnings:
+rr$errorsEmpty data.table (0 rows and 2 cols): iteration,msgrr$warningsEmpty data.table (0 rows and 2 cols): iteration,msgPerformance in each fold:
+rr$score(measures) task_id learner_id resampling_id iteration surv.graf surv.cindex.uno
+1: lung surv.bart cv 1 -0.312614598 0.5869665
+2: lung surv.bart cv 2 -0.103181391 0.5502903
+3: lung surv.bart cv 3 0.001448263 0.6178001
+4: lung surv.bart cv 4 -0.044161171 0.6157215
+5: lung surv.bart cv 5 -0.043129352 0.5688389
+Hidden columns: task, learner, resampling, predictionMean cross-validation performance:
+rr$aggregate(measures) surv.graf surv.cindex.uno
+ -0.1003276 0.5879235 We will choose two patients from the test set and plot their survival prediction posterior estimates.
+Let’s choose the patients with the worst and the best survival time:
+death_times = p$truth[,1]
+sort(death_times) [1] 3 5 5 6 6 6 7 8 8 8 8 10 10 10 12 12 12 13 15 16 17 18 27worst_indx = which(death_times == min(death_times))[1] # died first
+best_indx = which(death_times == max(death_times))[1] # died last
+
+patient_ids = c(worst_indx, best_indx)
+patient_ids # which patient IDs[1] 5 18death_times = death_times[patient_ids]
+death_times # 1st is worst, 2nd is best[1] 3 27Subset Arrdist to only the above 2 patients:
arrd = p$distr[patient_ids]
+arrdArrdist(2x31x50) We choose time points (in months) for the survival estimates:
+months = seq(1, 36) # 1 month - 3 yearsWe use the $distr interface and the $survival property to get survival probabilities from an Arrdist object as well as the quantile credible intervals (CIs). The median survival probabilities can be extracted as follows:
med = arrd$survival(months) # 'med' for median
+
+colnames(med) = paste0(patient_ids, "_med")
+med = as_tibble(med) %>% add_column(month = months)
+head(med)# A tibble: 6 × 3
+ `5_med` `18_med` month
+ <dbl> <dbl> <int>
+1 0.874 0.981 1
+2 0.767 0.962 2
+3 0.670 0.945 3
+4 0.569 0.927 4
+5 0.465 0.901 5
+6 0.366 0.869 6We can briefly verify model’s predictions: 1st patient survival probabilities on any month are lower (worst) compared to the 2nd patient.
+Note that subsetting an Arrdist (3d array) creates a Matdist (2d matrix), for example we can explicitly get the median survival probabilities:
matd_median = arrd[, 0.5] # median
+head(matd_median$survival(months)) # same as with `arrd` [,1] [,2]
+1 0.8741127 0.9808363
+2 0.7670382 0.9621618
+3 0.6701276 0.9450867
+4 0.5688809 0.9272284
+5 0.4647686 0.9007042
+6 0.3660939 0.8687270Using the mean posterior survival probabilities or the ones from the last posterior draw is also possible and can be done as follows:
matd_mean = arrd[, "mean"] # mean (if needed)
+head(matd_mean$survival(months)) [,1] [,2]
+1 0.8652006 0.9748463
+2 0.7533538 0.9521817
+3 0.6560050 0.9293229
+4 0.5623555 0.9051549
+5 0.4750038 0.8758896
+6 0.3815333 0.8360373matd_50draw = arrd[, 50] # the 50th posterior draw
+head(matd_50draw$survival(months)) [,1] [,2]
+1 0.9178342 0.9920982
+2 0.8424195 0.9842589
+3 0.7732014 0.9764815
+4 0.7096707 0.9687656
+5 0.6029119 0.9495583
+6 0.5122132 0.9307318To get the CIs we will subset the Arrdist using a quantile number (0-1), which extracts a Matdist based on the cdf. The survival function is 1 - cdf, so low and upper bounds are reversed:
low = arrd[, 0.975]$survival(months) # 2.5% bound
+high = arrd[, 0.025]$survival(months) # 97.5% bound
+colnames(low) = paste0(patient_ids, "_low")
+colnames(high) = paste0(patient_ids, "_high")
+low = as_tibble(low)
+high = as_tibble(high)The median posterior survival probabilities for the two patient of interest and the corresponding CI bounds in a tidy format are:
+surv_tbl =
+ bind_cols(low, med, high) %>%
+ pivot_longer(cols = !month, values_to = "surv",
+ names_to = c("patient_id", ".value"), names_sep = "_") %>%
+ relocate(patient_id)
+surv_tbl# A tibble: 72 × 5
+ patient_id month low med high
+ <chr> <int> <dbl> <dbl> <dbl>
+ 1 5 1 0.713 0.874 0.953
+ 2 18 1 0.929 0.981 0.996
+ 3 5 2 0.508 0.767 0.903
+ 4 18 2 0.863 0.962 0.991
+ 5 5 3 0.362 0.670 0.855
+ 6 18 3 0.801 0.945 0.985
+ 7 5 4 0.244 0.569 0.804
+ 8 18 4 0.734 0.927 0.977
+ 9 5 5 0.146 0.465 0.748
+10 18 5 0.654 0.901 0.969
+# ℹ 62 more rowsWe draw survival curves with the uncertainty for the survival probability quantified:
+my_colors = c("#E41A1C", "#4DAF4A")
+names(my_colors) = patient_ids
+
+surv_tbl %>%
+ ggplot(aes(x = month, y = med)) +
+ geom_step(aes(color = patient_id), linewidth = 1) +
+ xlab('Time (Months)') +
+ ylab('Survival Probability') +
+ geom_ribbon(aes(ymin = low, ymax = high, fill = patient_id),
+ alpha = 0.3, show.legend = F) +
+ geom_vline(xintercept = death_times[1], linetype = 'dashed', color = my_colors[1]) +
+ geom_vline(xintercept = death_times[2], linetype = 'dashed', color = my_colors[2]) +
+ theme_bw(base_size = 14) +
+ scale_color_manual(values = my_colors) +
+ scale_fill_manual(values = my_colors) +
+ guides(color = guide_legend(title = "Patient ID"))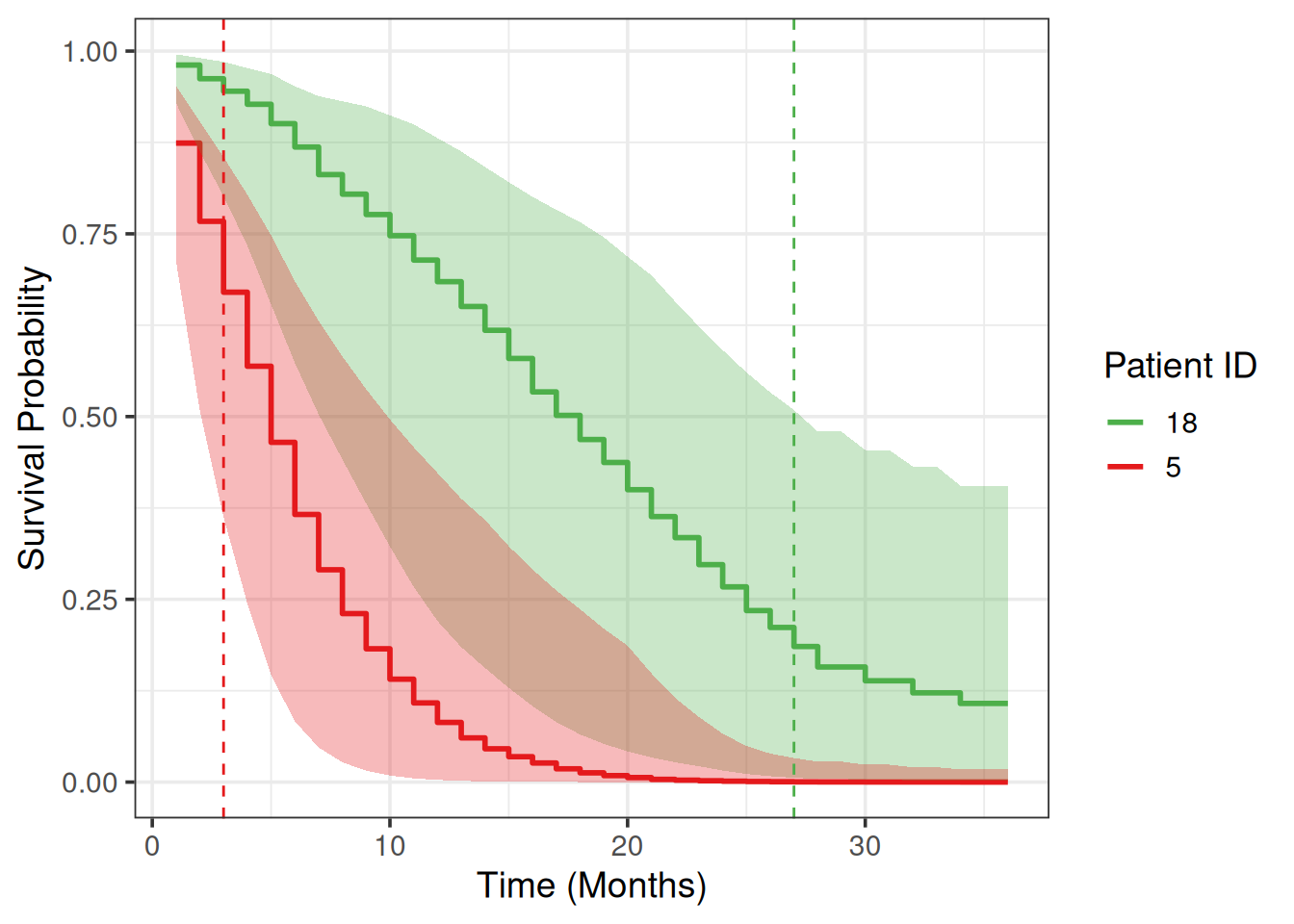
We will use a Partial Dependence Plot (PDP) (Friedman 2001) to visualize how much different are males vs females in terms of their average survival predictions across time.
+PDPs assume that features are independent. In our case we need to check that sex doesn’t correlate with any of the other features used for training the BART learner. Since sex is a categorical feature, we fit a linear model using as target variable every other feature in the data () and conduct an ANOVA (ANalysis Of VAriance) to get the variance explained or
. The square root of that value is the correlation measure we want.
# code from https://christophm.github.io/interpretable-ml-book/ale.html
+mycor = function(cnames, data) {
+ x.num = data[, cnames[1], with = FALSE][[1]]
+ x.cat = data[, cnames[2], with = FALSE][[1]]
+ # R^2 = Cor(X, Y)^2 in simple linear regression
+ sqrt(summary(lm(x.num ~ x.cat))$r.squared)
+}
+
+cnames = c("sex")
+combs = expand.grid(y = setdiff(colnames(d), "sex"), x = cnames)
+combs$cor = apply(combs, 1, mycor, data = task$data()) # use the train set
+combs y x cor
+1 time sex 0.12941337
+2 status sex 0.24343282
+3 age sex 0.12216709
+4 inst sex 0.07826337
+5 meal.cal sex 0.18389545
+6 pat.karno sex 0.04132443
+7 ph.ecog sex 0.02564987
+8 ph.karno sex 0.01702471
+9 wt.loss sex 0.13431983sex doesn’t correlate strongly with any other feature, so we can compute the PDP:
# create two datasets: one with males and one with females
+# all other features remain the same (use train data, 205 patients)
+d = task$data(rows = part$train) # `rows = part$test` to use the test set
+
+d$sex = 1
+task_males = as_task_surv(d, time = 'time', event = 'status', id = 'lung-males')
+d$sex = 0
+task_females = as_task_surv(d, time = 'time', event = 'status', id = 'lung-females')
+
+# make predictions
+p_males = learner$predict(task_males)
+p_females = learner$predict(task_females)
+
+# take the median posterior survival probability
+surv_males = p_males$distr$survival(months) # patients x times
+surv_females = p_females$distr$survival(months) # patients x times
+
+# tidy up data: average and quantiles across patients
+data_males =
+ apply(surv_males, 1, function(row) {
+ tibble(
+ low = quantile(row, probs = 0.025),
+ avg = mean(row),
+ high = quantile(row, probs = 0.975)
+ )
+ }) %>%
+ bind_rows() %>%
+ add_column(sex = 'male', month = months, .before = 1)
+
+data_females =
+ apply(surv_females, 1, function(row) {
+ tibble(
+ low = quantile(row, probs = 0.025),
+ avg = mean(row),
+ high = quantile(row, probs = 0.975)
+ )
+ }) %>%
+ bind_rows() %>%
+ add_column(sex = 'female', month = months, .before = 1)
+
+pdp_tbl = bind_rows(data_males, data_females)
+pdp_tbl# A tibble: 72 × 5
+ sex month low avg high
+ <chr> <int> <dbl> <dbl> <dbl>
+ 1 male 1 0.836 0.942 0.981
+ 2 male 2 0.704 0.889 0.963
+ 3 male 3 0.587 0.839 0.943
+ 4 male 4 0.488 0.788 0.924
+ 5 male 5 0.392 0.732 0.897
+ 6 male 6 0.304 0.663 0.860
+ 7 male 7 0.234 0.601 0.829
+ 8 male 8 0.172 0.550 0.799
+ 9 male 9 0.130 0.503 0.766
+10 male 10 0.0945 0.455 0.733
+# ℹ 62 more rowsmy_colors = c("#E41A1C", "#4DAF4A")
+names(my_colors) = c('male', 'female')
+
+pdp_tbl %>%
+ ggplot(aes(x = month, y = avg)) +
+ geom_step(aes(color = sex), linewidth = 1) +
+ xlab('Time (Months)') +
+ ylab('Survival Probability') +
+ geom_ribbon(aes(ymin = low, ymax = high, fill = sex), alpha = 0.2, show.legend = F) +
+ theme_bw(base_size = 14) +
+ scale_color_manual(values = my_colors) +
+ scale_fill_manual(values = my_colors)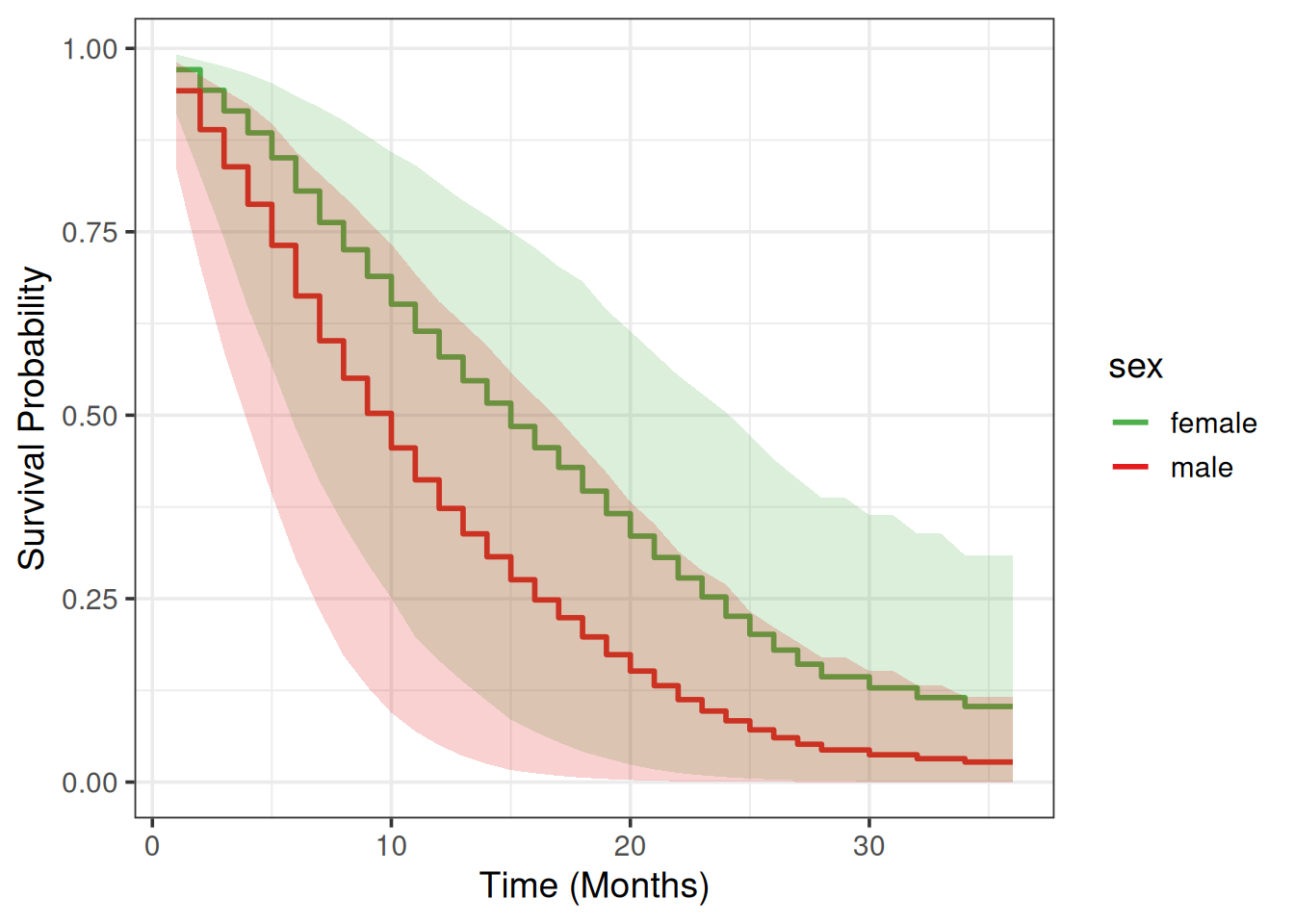
Working with spatial data in R requires a lot of data wrangling e.g. reading from different file formats, converting between spatial formats, creating tables from point layers, and predicting spatial raster images. The goal of mlr3spatial is to simplify these workflows within the mlr3 ecosystem. As a practical example, we will perform a land cover classification for the city of Leipzig, Germany. Figure 1 illustrates the typical workflow for this type of task: Load the training data, create a spatial task, train a learner with it, and predict the final raster image.
+%%{ init: { 'flowchart': { 'curve': 'bump' } } }%%
+
+flowchart LR
+ subgraph files[Files]
+ vector[Vector]
+ raster[Raster]
+ end
+ subgraph load[Load Data]
+ sf
+ terra
+ end
+ vector --> sf
+ raster --> terra
+ subgraph train_model[Train Model]
+ task[Task]
+ learner[Learner]
+ end
+ terra --> prediction_raster
+ task --> learner
+ sf --> task
+ subgraph predict[Spatial Prediction]
+ prediction_raster[Raster Image]
+ end
+ learner --> prediction_raster
+
+We assume that you are familiar with the mlr3 ecosystem and know the basic concepts of remote sensing. If not, we recommend reading the mlr3book first. If you are interested in spatial resampling, check out the book chapter on spatial analysis.
+ +Land cover is the physical material or vegetation that covers the surface of the earth, including both natural and human-made features. Understanding land cover patterns and changes over time is critical for addressing global environmental challenges, such as climate change, land degradation, and loss of biodiversity. Land cover classification is the process of assigning land cover classes to pixels in a raster image. With mlr3spatial, we can easily perform a land cover classification within the mlr3 ecosystem.
+Before we can start the land cover classification, we need to load the necessary packages. The mlr3spatial package relies on terra for processing raster data and sf for vector data. These widely used packages read all common raster and vector formats. Additionally, the stars and raster packages are supported.
+library(mlr3verse)
+library(mlr3spatial)
+library(terra, exclude = "resample")
+library(sf)We will work with a Sentinel-2 scene of the city of Leipzig which consists of 7 bands with a 10 or 20m spatial resolution and an NDVI band. The data is included in the mlr3spatial package. We use the terra::rast() to load the TIFF raster file.
leipzig_raster = rast(system.file("extdata", "leipzig_raster.tif", package = "mlr3spatial"))
+leipzig_rasterclass : SpatRaster
+dimensions : 206, 154, 8 (nrow, ncol, nlyr)
+resolution : 10, 10 (x, y)
+extent : 731810, 733350, 5692030, 5694090 (xmin, xmax, ymin, ymax)
+coord. ref. : WGS 84 / UTM zone 32N (EPSG:32632)
+source : leipzig_raster.tif
+names : b02, b03, b04, b06, b07, b08, ...
+min values : 846, 645, 366, 375, 401, 374, ...
+max values : 4705, 4880, 5451, 4330, 5162, 5749, ... The training data is a GeoPackage point layer with land cover labels and spectral features. We load the file and create a simple feature point layer.
leipzig_vector = read_sf(system.file("extdata", "leipzig_points.gpkg", package = "mlr3spatial"), stringsAsFactors = TRUE)
+leipzig_vectorSimple feature collection with 97 features and 9 fields
+Geometry type: POINT
+Dimension: XY
+Bounding box: xmin: 731930.5 ymin: 5692136 xmax: 733220.3 ymax: 5693968
+Projected CRS: WGS 84 / UTM zone 32N
+# A tibble: 97 × 10
+ b02 b03 b04 b06 b07 b08 b11 ndvi land_cover geom
+ <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <POINT [m]>
+ 1 903 772 426 2998 4240 4029 1816 0.809 forest (732480.1 5693957)
+ 2 1270 1256 1081 1998 2493 2957 2073 0.465 urban (732217.4 5692769)
+ 3 1033 996 777 2117 2748 2799 1595 0.565 urban (732737.2 5692469)
+ 4 962 773 500 465 505 396 153 -0.116 water (733169.3 5692777)
+ 5 1576 1527 1626 1715 1745 1768 1980 0.0418 urban (732202.2 5692644)
+ 6 1125 1185 920 3058 3818 3758 2682 0.607 pasture (732153 5693059)
+ 7 880 746 424 2502 3500 3397 1469 0.778 forest (731937.9 5693722)
+ 8 1332 1251 1385 1663 1799 1640 1910 0.0843 urban (732416.2 5692324)
+ 9 940 741 475 452 515 400 139 -0.0857 water (732933.7 5693344)
+10 902 802 454 2764 3821 3666 1567 0.780 forest (732411.3 5693352)
+# ℹ 87 more rowsWe plot both layers to get an overview of the data. The training points are located in the districts of Lindenau and Zentrum West.
+library(ggplot2)
+library(tidyterra, exclude = "filter")
+
+ggplot() +
+ geom_spatraster_rgb(data = leipzig_raster, r = 3, g = 2, b = 1, max_col_value = 5451) +
+ geom_spatvector(data = leipzig_vector, aes(color = land_cover)) +
+ scale_color_viridis_d(name = "Land cover", labels = c("Forest", "Pastures", "Urban", "Water")) +
+ theme_minimal()
The as_task_classif_st() function directly creates a spatial task from the point layer. This makes it unnecessary to transform the point layer to a data.frame with coordinates. Spatial tasks additionally store the coordinates of the training points. The coordinates are useful when estimating the performance of the model with spatial resampling.
task = as_task_classif_st(leipzig_vector, target = "land_cover")
+task<TaskClassifST:leipzig_vector> (97 x 9)
+* Target: land_cover
+* Properties: multiclass
+* Features (8):
+ - dbl (8): b02, b03, b04, b06, b07, b08, b11, ndvi
+* Coordinates:
+ X Y
+ 1: 732480.1 5693957
+ 2: 732217.4 5692769
+ 3: 732737.2 5692469
+ 4: 733169.3 5692777
+ 5: 732202.2 5692644
+---
+93: 733018.7 5692342
+94: 732551.4 5692887
+95: 732520.4 5692589
+96: 732542.2 5692204
+97: 732437.8 5692300Now we can train a model with the task. We use a simple decision tree learner from the rpart package. The "classif_st" task is a specialization of the "classif" task and therefore works with all "classif" learners.
learner = lrn("classif.rpart")
+learner$train(task)To get a complete land cover classification of Leipzig, we have to predict on each pixel and return a raster image with these predictions. The $predict() method of the learner only works for tabular data. To predict a raster image, we use the predict_spatial() function.
# predict land cover map
+land_cover = predict_spatial(leipzig_raster, learner)ggplot() +
+ geom_spatraster(data = land_cover) +
+ scale_fill_viridis_d(name = "Land cover", labels = c("Forest", "Pastures", "Urban", "Water")) +
+ theme_minimal()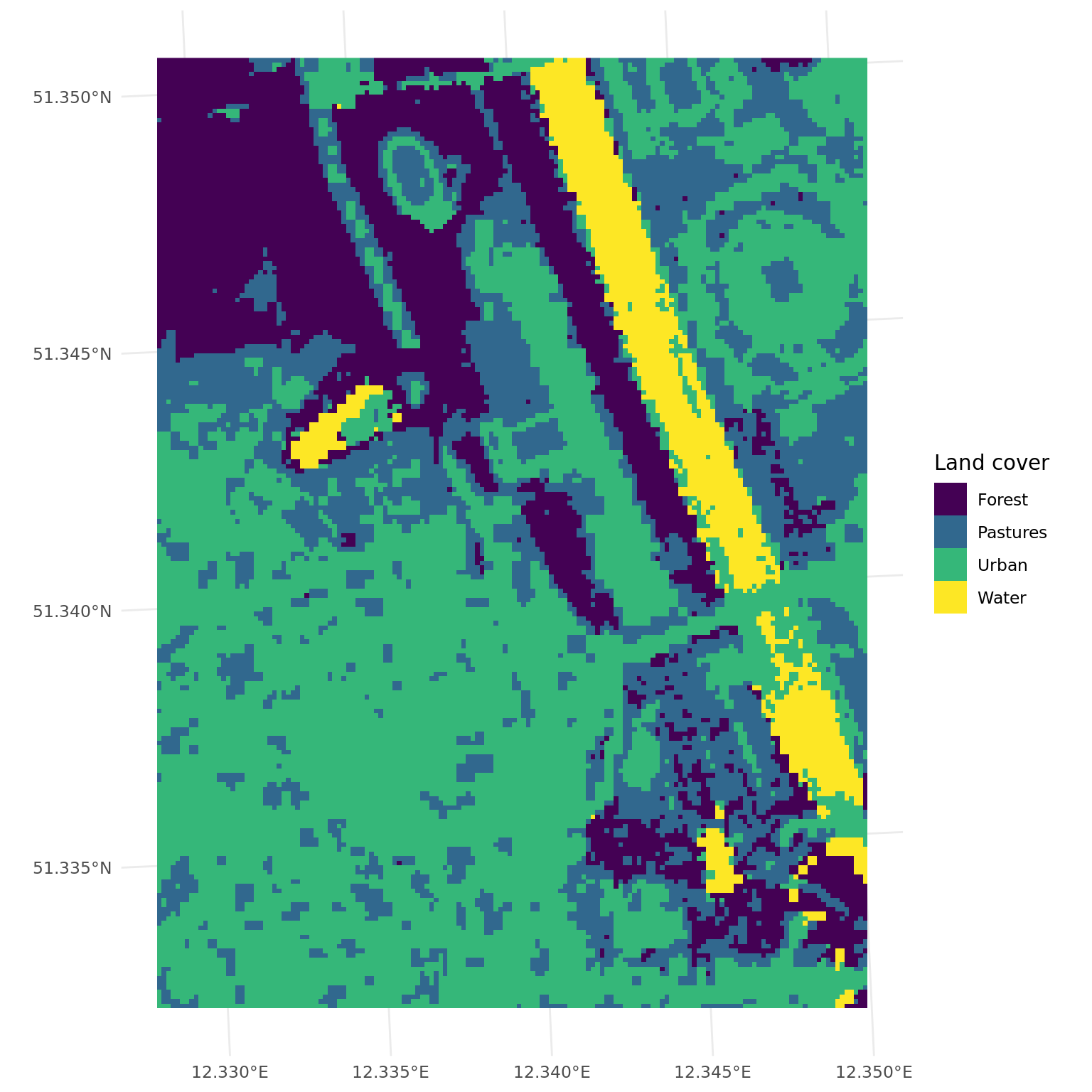
Working with spatial data in R is very easy with the mlr3spatial package. You can quickly train a model with a point layer and predict a raster image. The mlr3spatial package is still in development and we are looking forward to your feedback and contributions.
+ + +Feature selection is the process of finding an optimal subset of features in order to improve the performance, interpretability and robustness of machine learning algorithms. In this article, we introduce the wrapper feature selection method Recursive Feature Elimination. Wrapper methods iteratively select features that optimize a performance measure. As an example, we will search for the optimal set of features for a gradient boosting machine and support vector machine on the Sonar data set. We assume that you are already familiar with the basic building blocks of the mlr3 ecosystem. If you are new to feature selection, we recommend reading the feature selection chapter of the mlr3book first.
Recursive Feature Elimination (RFE) is a widely used feature selection method for high-dimensional data sets. The idea is to iteratively remove the least predictive feature from a model until the desired number of features is reached. This feature is determined by the built-in feature importance method of the model. Currently, RFE works with support vector machines (SVM), decision tree algorithms and gradient boosting machines (GBM). Supported learners are tagged with the "importance" property. For a full list of supported learners, see the learner page on the mlr-org website and search for "importance".
Guyon et al. (2002) developed the RFE algorithm for SVMs (SVM-RFE) to select informative genes in cancer classification. The importance of the features is given by the weight vector of a linear support vector machine. This method was later extended to other machine learning algorithms. The only requirement is that the models can internally measure the feature importance. The random forest algorithm offers multiple options for measuring feature importance. The commonly used methods are the mean decrease in accuracy (MDA) and the mean decrease in impurity (MDI). The MDA measures the decrease in accuracy for a feature if it was randomly permuted in the out-of-bag sample. The MDI is the total reduction in node impurity when the feature is used for splitting. Gradient boosting algorithms like XGBoost, LightGBM and GBM use similar methods to measure the importance of the features.
Resampling strategies can be combined with the algorithm in different ways. The frameworks scikit-learn (Pedregosa et al. 2011) and caret (Kuhn 2008) implement a variant called recursive feature elimination with cross-validation (RFE-CV) that estimates the optimal number of features with cross-validation first. Then one more RFE is carried out on the complete dataset with the optimal number of features as the final feature set size. The RFE implementation in mlr3 can rank and aggregate importance scores across resampling iterations. We will explore both variants in more detail below.
+mlr3fselect is the feature selection package of the mlr3 ecosystem. It implements the RFE and RFE-CV algorithm. We load all packages of the ecosystem with the mlr3verse package.
library(mlr3verse)We retrieve the RFE optimizer with the fs() function.
optimizer = fs("rfe",
+ n_features = 1,
+ feature_number = 1,
+ aggregation = "rank")The algorithm has multiple control parameters. The optimizer stops when the number of features equals n_features. The parameters feature_number, feature_fraction and subset_size determine the number of features that are removed in each iteration. The feature_number option removes a fixed number of features in each iteration, whereas feature_fraction removes a fraction of the features. The subset_size argument is a vector that specifies exactly how many features are removed in each iteration. The parameters are mutually exclusive and the default is feature_fraction = 0.5. Usually, RFE fits a new model in each resampling iteration and calculates the feature importance again. We can deactivate this behavior by setting recursive = FALSE. The selection of feature subsets in all iterations is then based solely on the importance scores of the first model trained with all features. When running an RFE with a resampling strategy like cross-validation, multiple models and importance scores are generated. The aggregation parameter determines how the importance scores are aggregated. The option "rank" ranks the importance scores in each iteration and then averages the ranks of the features. The feature with the lowest average rank is removed. The option "mean" averages the importance scores of the features across the iterations. The "mean" should only be used if the learner’s importance scores can be reasonably averaged.
The objective of the Sonar data set is to predict whether a sonar signal bounced off a metal cylinder or a rock. The data set includes 60 numerical features (see Figure 1).
task = tsk("sonar")library(ggplot2)
+library(data.table)
+
+data = melt(as.data.table(task), id.vars = task$target_names, measure.vars = task$feature_names)
+data = data[c("V1", "V10", "V11", "V12", "V13", "V14"), , on = "variable"]
+
+ggplot(data, aes(x = value, fill = Class)) +
+ geom_density(alpha = 0.5) +
+ facet_wrap(~ variable, ncol = 6, scales = "free") +
+ scale_fill_viridis_d(end = 0.8) +
+ theme_minimal() +
+ theme(axis.title.x = element_blank())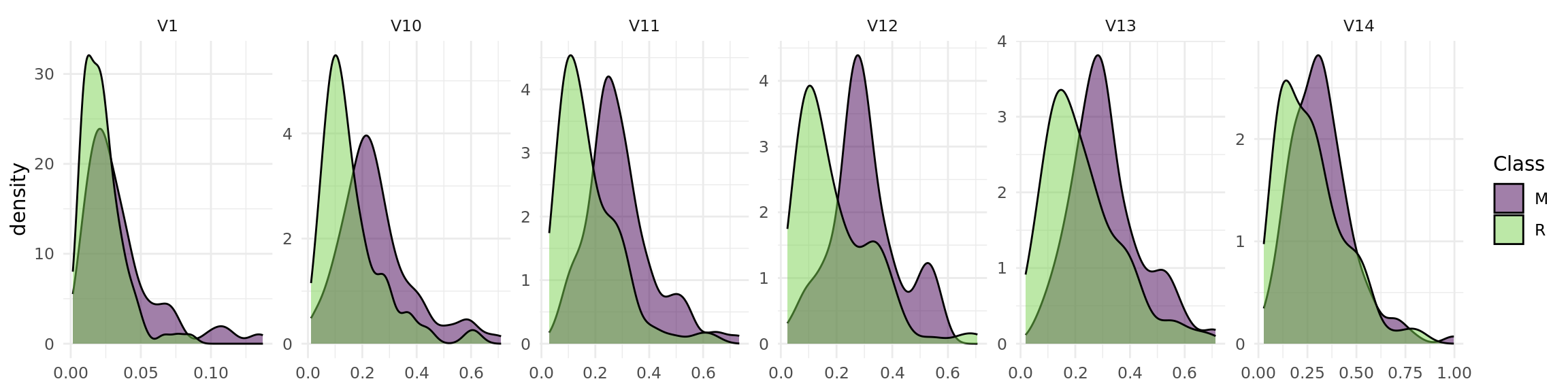 +
+We start with the GBM learner and set the predict type to "prob" to obtain class probabilities.
learner = lrn("classif.gbm",
+ distribution = "bernoulli",
+ predict_type = "prob")Now we define the feature selection problem by using the fsi() function that constructs an FSelectInstanceSingleCrit. In addition to the task and learner, we have to select a resampling strategy and performance measure to determine how the performance of a feature subset is evaluated. We pass the "none" terminator because the n_features parameter of the optimizer determines when the feature selection stops.
instance = fsi(
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 6),
+ measures = msr("classif.auc"),
+ terminator = trm("none"))We are now ready to start the RFE. To do this, we simply pass the instance to the $optimize() method of the optimizer.
optimizer$optimize(instance)The optimizer saves the best feature set and the corresponding estimated performance in instance$result.
Figure 2 shows the optimization path of the feature selection. We observe that the performance increases first as the number of features decreases. As soon as informative features are removed, the performance drops.
+library(viridisLite)
+library(mlr3misc)
+
+data = as.data.table(instance$archive)
+data[, n:= map_int(importance, length)]
+
+ggplot(data, aes(x = n, y = classif.auc)) +
+ geom_line(
+ color = viridis(1, begin = 0.5),
+ linewidth = 1) +
+ geom_point(
+ fill = viridis(1, begin = 0.5),
+ shape = 21,
+ size = 3,
+ stroke = 0.5,
+ alpha = 0.8) +
+ xlab("Number of Features") +
+ scale_x_reverse() +
+ theme_minimal()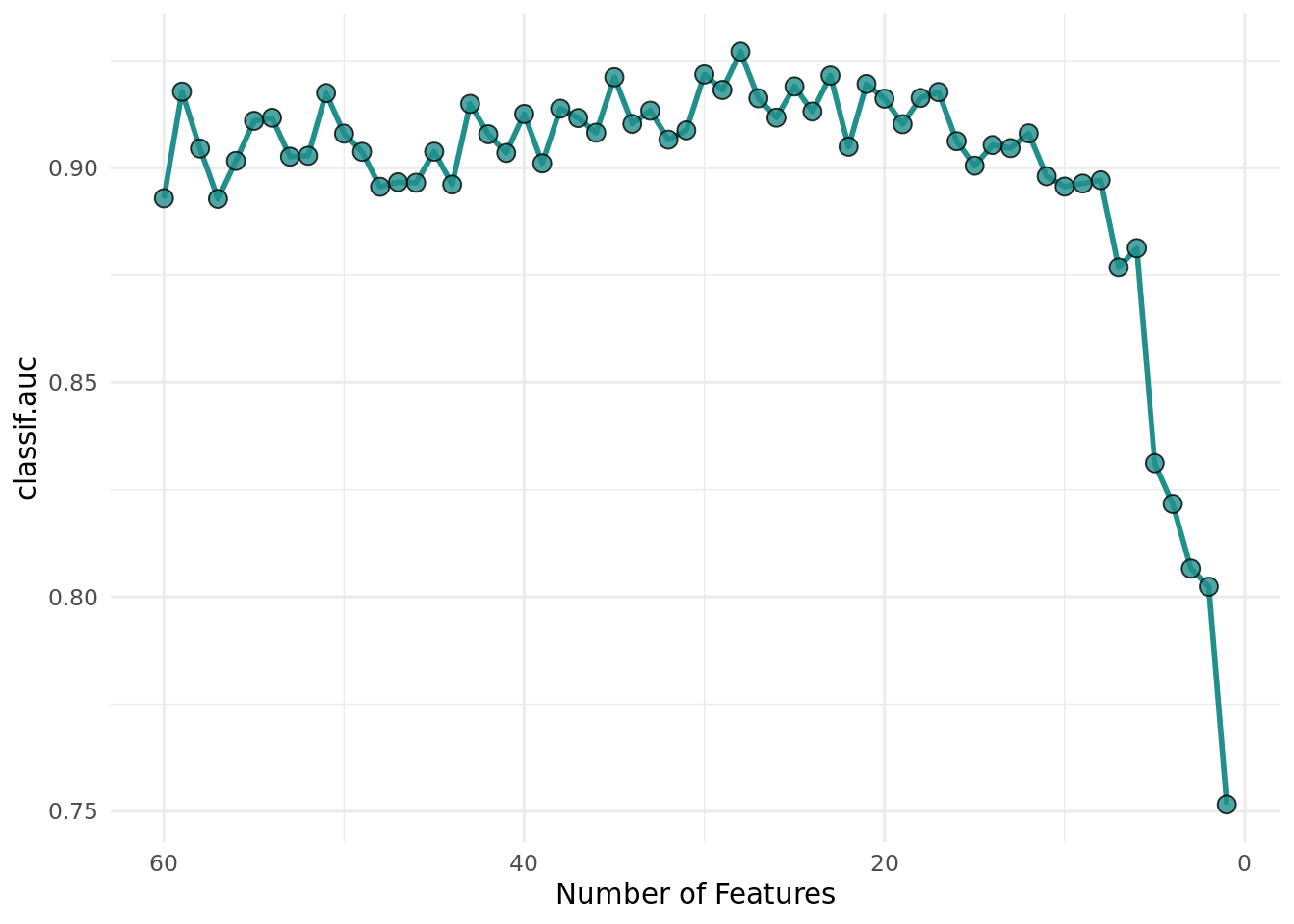 +
+The importance scores of the feature sets are recorded in the archive.
+as.data.table(instance$archive)[, list(features, classif.auc, importance)] features classif.auc importance
+ 1: V1,V10,V11,V12,V13,V14,... 0.8929304 58.83333,58.83333,54.50000,54.00000,53.33333,52.50000,...
+ 2: V1,V10,V11,V12,V13,V15,... 0.9177811 57.33333,56.00000,54.00000,53.66667,50.50000,50.00000,...
+ 3: V1,V10,V11,V12,V13,V15,... 0.9045253 54.83333,54.66667,54.66667,53.00000,51.83333,51.33333,...
+ 4: V1,V10,V11,V12,V13,V15,... 0.8927833 56.00000,55.83333,53.00000,52.00000,50.16667,50.00000,...
+ 5: V1,V10,V11,V12,V13,V15,... 0.9016274 55.50000,53.50000,51.33333,50.00000,49.00000,48.50000,...
+---
+56: V11,V12,V16,V48,V9 0.8311625 4.166667,3.333333,2.833333,2.500000,2.166667
+57: V11,V12,V16,V9 0.8216772 3.833333,2.666667,2.000000,1.500000
+58: V11,V12,V16 0.8065807 2.833333,1.833333,1.333333
+59: V11,V12 0.8023780 1.833333,1.166667
+60: V11 0.7515904 1Now we will select the optimal feature set for an SVM with a linear kernel. The importance scores are the weights of the model.
+learner = lrn("classif.svm",
+ type = "C-classification",
+ kernel = "linear",
+ predict_type = "prob")The SVM learner does not support the calculation of importance scores at first. The reason is that importance scores cannot be determined with all kernels. This can be seen by the missing "importance" property.
learner$properties[1] "multiclass" "twoclass" Using the "mlr3fselect.svm_rfe" callback however makes it possible to use a linear SVM with the RFE optimizer. The callback adds the $importance() method internally to the learner. We load the callback with the clbk() function and pass it as the "callback" argument to fsi().
instance = fsi(
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 6),
+ measures = msr("classif.auc"),
+ terminator = trm("none"),
+ callback = clbk("mlr3fselect.svm_rfe"))We start the feature selection.
+optimizer$optimize(instance)Figure 3 shows the average performance of the SVMs depending on the number of features. We can see that the performance increases significantly with a reduced feature set.
+library(viridisLite)
+library(mlr3misc)
+
+data = as.data.table(instance$archive)
+data[, n:= map_int(importance, length)]
+
+ggplot(data, aes(x = n, y = classif.auc)) +
+ geom_line(
+ color = viridis(1, begin = 0.5),
+ linewidth = 1) +
+ geom_point(
+ fill = viridis(1, begin = 0.5),
+ shape = 21,
+ size = 3,
+ stroke = 0.5,
+ alpha = 0.8) +
+ xlab("Number of Features") +
+ scale_x_reverse() +
+ theme_minimal()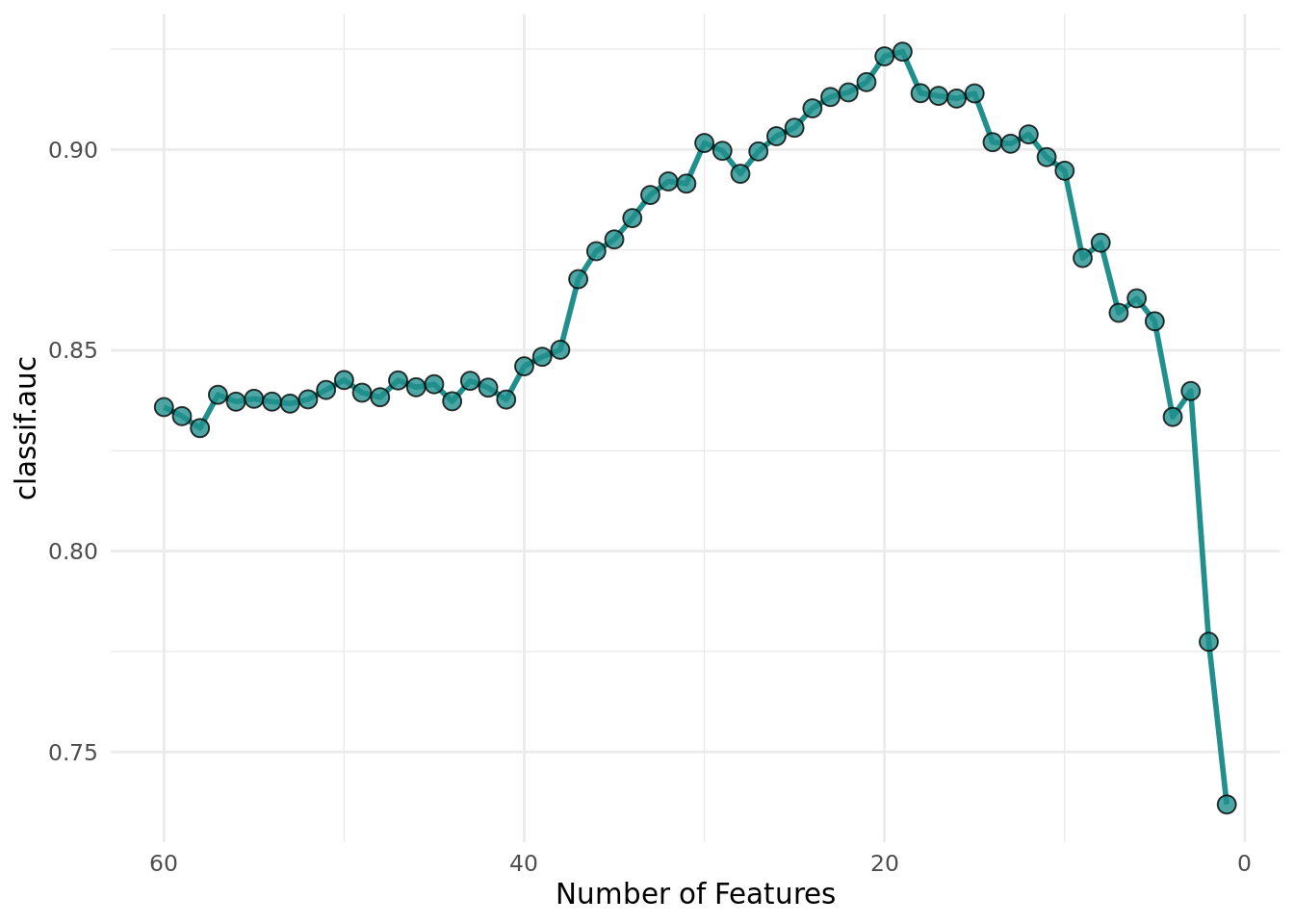 +
+For datasets with a lot of features, it is more efficient to remove several features per iteration. We show an example where 25% of the features are removed in each iteration.
+optimizer = fs("rfe", n_features = 1, feature_fraction = 0.75)
+
+instance = fsi(
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 6),
+ measures = msr("classif.auc"),
+ terminator = trm("none"),
+ callback = clbk("mlr3fselect.svm_rfe"))
+
+optimizer$optimize(instance)Figure 4 shows a similar optimization curve as Figure 3 but with fewer evaluated feature sets.
+library(viridisLite)
+library(mlr3misc)
+
+data = as.data.table(instance$archive)
+data[, n:= map_int(importance, length)]
+
+ggplot(data, aes(x = n, y = classif.auc)) +
+ geom_line(
+ color = viridis(1, begin = 0.5),
+ linewidth = 1) +
+ geom_point(
+ fill = viridis(1, begin = 0.5),
+ shape = 21,
+ size = 3,
+ stroke = 0.5,
+ alpha = 0.8) +
+ xlab("Number of Features") +
+ scale_x_reverse() +
+ theme_minimal()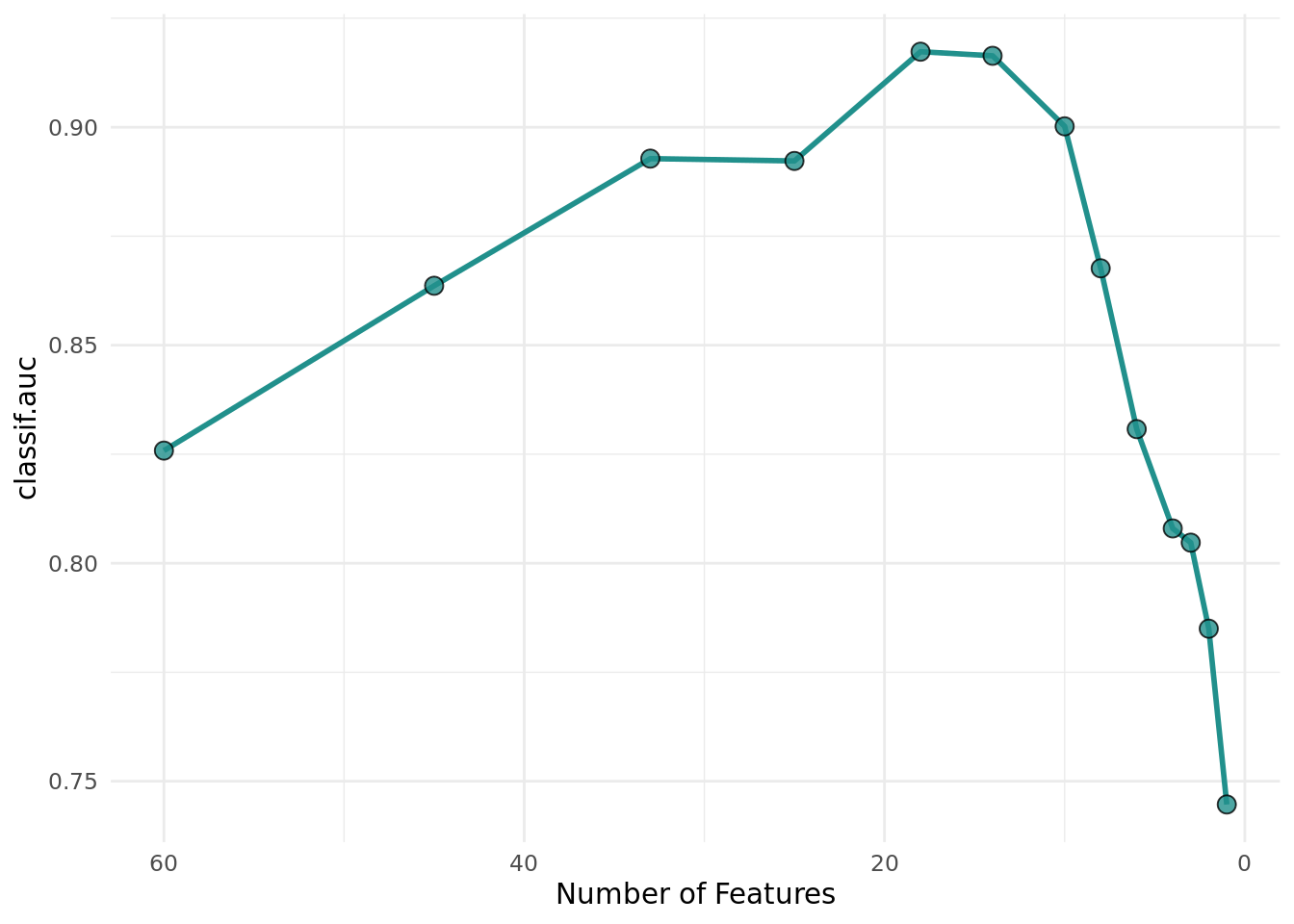 +
+RFE-CV estimates the optimal number of features before selecting a feature set. For this, an RFE is run in each resampling iteration and the number of features with the best mean performance is selected (see Figure 5). Then one more RFE is carried out on the complete dataset with the optimal number of features as the final feature set size.
+%%{ init: { 'flowchart': { 'curve': 'bump' } } }%%
+flowchart TB
+ cross-validation[3-Fold Cross-Validation]
+ cross-validation-->rfe-1
+ cross-validation-->rfe-2
+ cross-validation-->rfe-3
+ subgraph rfe-1[RFE 1]
+ direction TB
+ f14[4 Features]
+ f13[3 Features]
+ f12[2 Features]
+ f11[1 Features]
+ f14-->f13-->f12-->f11
+ style f13 fill:#ccea84
+ end
+ subgraph rfe-2[RFE 2]
+ direction TB
+ f24[4 Features]
+ f23[3 Features]
+ f22[2 Features]
+ f21[1 Features]
+ f24-->f23-->f22-->f21
+ style f23 fill:#ccea84
+ end
+ subgraph rfe-3[RFE 3]
+ direction TB
+ f34[4 Features]
+ f33[3 Features]
+ f32[2 Features]
+ f31[1 Features]
+ f34-->f33-->f32-->f31
+ style f33 fill:#ccea84
+ end
+ all_obs[All Observations]
+ rfe-1-->all_obs
+ rfe-2-->all_obs
+ rfe-3-->all_obs
+ all_obs --> rfe
+ subgraph rfe[RFE]
+ direction TB
+ f54[4 Features]
+ f53[3 Features]
+ f54-->f53
+ style f53 fill:#8e6698
+ end
+
+We retrieve the RFE-CV optimizer. RFE-CV has almost the same control parameters as the RFE optimizer. The only difference is that no aggregation is needed.
optimizer = fs("rfecv",
+ n_features = 1,
+ feature_number = 1)The chosen resampling strategy is used to estimate the optimal number of features. The 6-fold cross-validation results in 6 RFE runs. You can choose any other resampling strategy with multiple iterations. Let’s start the feature selection.
+learner = lrn("classif.svm",
+ type = "C-classification",
+ kernel = "linear",
+ predict_type = "prob")
+
+instance = fsi(
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 6),
+ measures = msr("classif.auc"),
+ terminator = trm("none"),
+ callback = clbk("mlr3fselect.svm_rfe"))
+
+optimizer$optimize(instance)The performance of the optimal feature set is calculated on the complete data set and should not be reported as the performance of the final model. Estimate the performance of the final model with nested resampling.
+We visualize the selection of the optimal number of features. Each point is the mean performance of the number of features. We achieved the best performance with 19 features.
+library(ggplot2)
+library(viridisLite)
+library(mlr3misc)
+
+data = as.data.table(instance$archive)[!is.na(iteration), ]
+aggr = data[, list("y" = mean(unlist(.SD))), by = "batch_nr", .SDcols = "classif.auc"]
+aggr[, batch_nr := 61 - batch_nr]
+
+data[, n:= map_int(importance, length)]
+
+ggplot(aggr, aes(x = batch_nr, y = y)) +
+ geom_line(
+ color = viridis(1, begin = 0.5),
+ linewidth = 1) +
+ geom_point(
+ fill = viridis(1, begin = 0.5),
+ shape = 21,
+ size = 3,
+ stroke = 0.5,
+ alpha = 0.8) +
+ geom_vline(
+ xintercept = aggr[y == max(y)]$batch_nr,
+ colour = viridis(1, begin = 0.33),
+ linetype = 3
+ ) +
+ xlab("Number of Features") +
+ ylab("Mean AUC") +
+ scale_x_reverse() +
+ theme_minimal()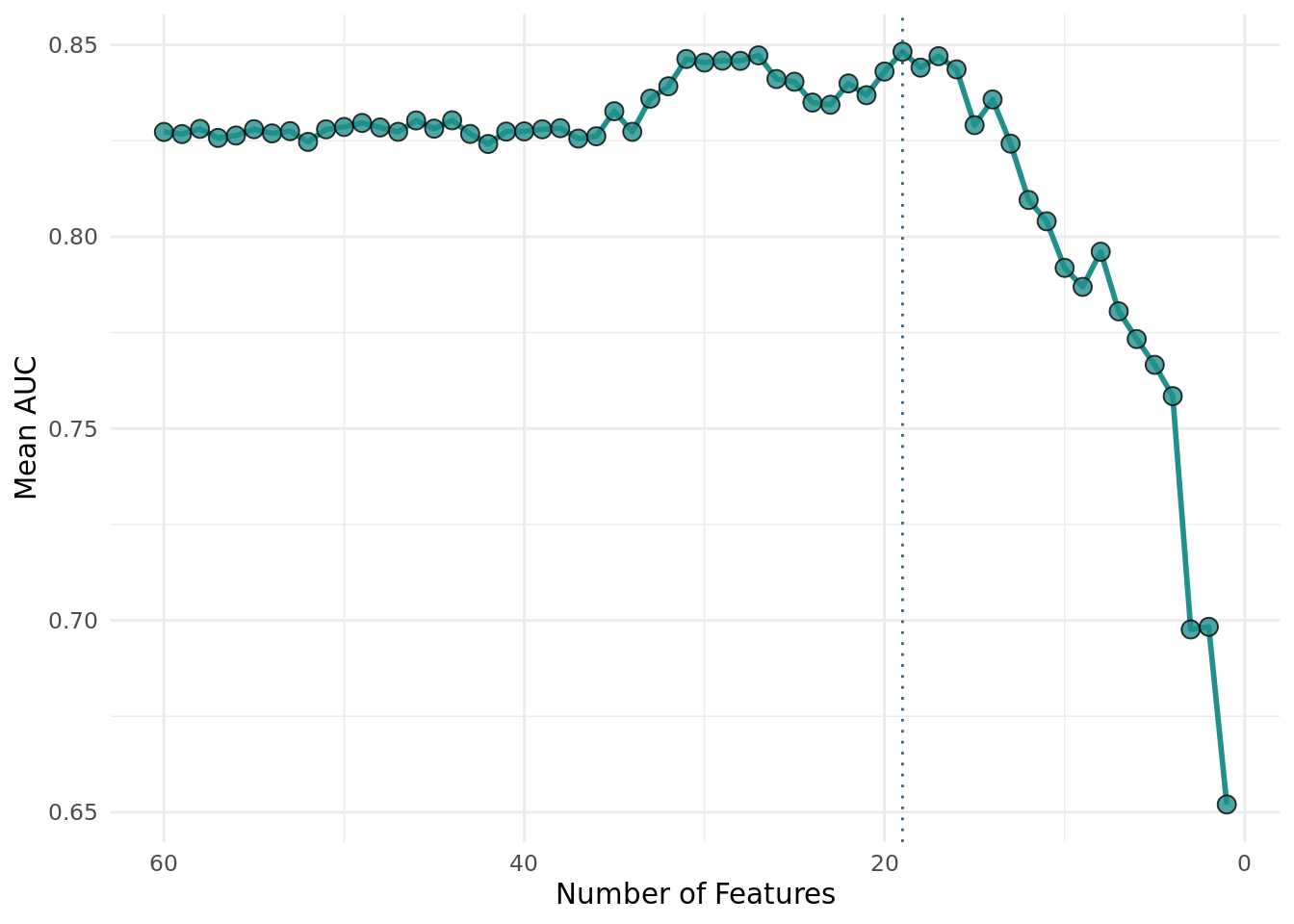 +
+The archive contains the extra column "iteration" that indicates in which resampling iteration the feature set was evaluated. The feature subsets of the final RFE run have no value in the "iteration" column because they were evaluated on the complete data set.
as.data.table(instance$archive)[, list(features, classif.auc, iteration, importance)] features classif.auc iteration importance
+ 1: V1,V10,V11,V12,V13,V14,... 0.8782895 1 2.864018,1.532774,1.408485,1.399930,1.326165,1.167745,...
+ 2: V1,V10,V11,V12,V13,V14,... 0.7026144 2 2.056442,1.706077,1.258703,1.191762,1.190752,1.178514,...
+ 3: V1,V10,V11,V12,V13,V14,... 0.8790850 3 1.950412,1.887710,1.820891,1.616219,1.231928,1.138675,...
+ 4: V1,V10,V11,V12,V13,V14,... 0.8125000 4 2.6958580,1.5623759,1.4990138,1.3902109,0.9385757,0.9232132,...
+ 5: V1,V10,V11,V12,V13,V14,... 0.8807018 5 2.487483,1.470778,1.356517,1.033764,0.635383,0.575074,...
+ ---
+398: V1,V11,V12,V16,V23,V3,... 0.9605275 NA 2.0089739,1.1047492,1.0011253,0.6602411,0.6015470,0.5431803,...
+399: V1,V12,V16,V23,V3,V30,... 0.9595988 NA 1.8337471,1.1937962,0.9853467,0.7751384,0.7296726,0.6222569,...
+400: V1,V12,V16,V23,V3,V30,... 0.9589486 NA 1.8824952,1.2468164,1.0106654,0.8090618,0.6983925,0.6568389,...
+401: V1,V12,V16,V23,V3,V30,... 0.9559766 NA 2.3872902,0.9094028,0.8809098,0.8277941,0.7841591,0.7792772,...
+402: V1,V12,V16,V23,V3,V30,... 0.9521687 NA 1.9485133,1.1482257,1.1098823,0.9591012,0.8234140,0.8118616,...The learner we use to make predictions on new data is called the final model. The final model is trained with the optimal feature set on the full data set. The optimal set consists of 19 features and is stored in instance$result_feature_set. We subset the task to the optimal feature set and train the learner.
task$select(instance$result_feature_set)
+learner$train(task)The trained model can now be used to predict new, external data.
+The RFE algorithm is a valuable feature selection method, especially for high-dimensional datasets with only a few observations. The numerous settings of the algorithm in mlr3 make it possible to apply it to many datasets and learners. If you want to know more about feature selection in general, we recommend having a look at our book.
+ + + +Feature selection is the process of finding an optimal set of features to improve the performance, interpretability and robustness of machine learning algorithms. In this article, we introduce the Shadow Variable Search algorithm which is a wrapper method for feature selection. Wrapper methods iteratively add features to the model that optimize a performance measure. As an example, we will search for the optimal set of features for a support vector machine on the Pima Indian Diabetes data set. We assume that you are already familiar with the basic building blocks of the mlr3 ecosystem. If you are new to feature selection, we recommend reading the feature selection chapter of the mlr3book first. Some knowledge about mlr3pipelines is beneficial but not necessary to understand the example.
Adding shadow variables to a data set is a well-known method in machine learning (Wu, Boos, and Stefanski 2007; Thomas et al. 2017). The idea is to add permutated copies of the original features to the data set. These permutated copies are called shadow variables or pseudovariables and the permutation breaks any relationship with the target variable, making them useless for prediction. The subsequent search is similar to the sequential forward selection algorithm, where one new feature is added in each iteration of the algorithm. This new feature is selected as the one that improves the performance of the model the most. This selection is computationally expensive, as one model for each of the not yet included features has to be trained. The difference between shadow variable search and sequential forward selection is that the former uses the selection of a shadow variable as the termination criterion. Selecting a shadow variable means that the best improvement is achieved by adding a feature that is unrelated to the target variable. Consequently, the variables not yet selected are most likely also correlated to the target variable only by chance. Therefore, only the previously selected features have a true influence on the target variable.
+mlr3fselect is the feature selection package of the mlr3 ecosystem. It implements the shadow variable search algorithm. We load all packages of the ecosystem with the mlr3verse package.
library(mlr3verse)We retrieve the shadow variable search optimizer with the fs() function. The algorithm has no control parameters.
optimizer = fs("shadow_variable_search")The objective of the Pima Indian Diabetes data set is to predict whether a person has diabetes or not. The data set includes 768 patients with 8 measurements (see Figure 1).
task = tsk("pima")library(ggplot2)
+library(data.table)
+
+data = melt(as.data.table(task), id.vars = task$target_names, measure.vars = task$feature_names)
+
+ggplot(data, aes(x = value, fill = diabetes)) +
+ geom_density(alpha = 0.5) +
+ facet_wrap(~ variable, ncol = 8, scales = "free") +
+ scale_fill_viridis_d(end = 0.8) +
+ theme_minimal() +
+ theme(axis.title.x = element_blank())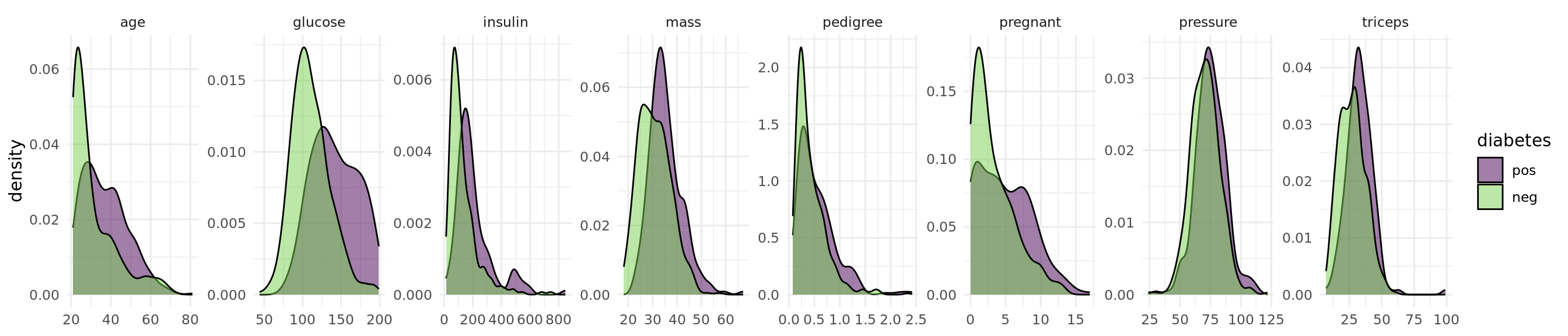 +
+The data set contains missing values.
+task$missings()diabetes age glucose insulin mass pedigree pregnant pressure triceps
+ 0 0 5 374 11 0 0 35 227 Support vector machines cannot handle missing values. We impute the missing values with the histogram imputation method.
learner = po("imputehist") %>>% lrn("classif.svm", predict_type = "prob")Now we define the feature selection problem by using the fsi() function that constructs an FSelectInstanceSingleCrit. In addition to the task and learner, we have to select a resampling strategy and performance measure to determine how the performance of a feature subset is evaluated. We pass the "none" terminator because the shadow variable search algorithm terminates by itself.
instance = fsi(
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 3),
+ measures = msr("classif.auc"),
+ terminator = trm("none")
+)We are now ready to start the shadow variable search. To do this, we simply pass the instance to the $optimize() method of the optimizer.
optimizer$optimize(instance) age glucose insulin mass pedigree pregnant pressure triceps features classif.auc
+1: TRUE TRUE FALSE TRUE TRUE FALSE FALSE FALSE age,glucose,mass,pedigree 0.835165The optimizer returns the best feature set and the corresponding estimated performance.
+Figure 2 shows the optimization path of the feature selection. The feature glucose was selected first and in the following iterations age, mass and pedigree. Then a shadow variable was selected and the feature selection was terminated.
+library(data.table)
+library(ggplot2)
+library(mlr3misc)
+library(viridisLite)
+
+data = as.data.table(instance$archive)[order(-classif.auc), head(.SD, 1), by = batch_nr][order(batch_nr)]
+data[, features := map_chr(features, str_collapse)]
+data[, batch_nr := as.character(batch_nr)]
+
+ggplot(data, aes(x = batch_nr, y = classif.auc)) +
+ geom_bar(
+ stat = "identity",
+ width = 0.5,
+ fill = viridis(1, begin = 0.5),
+ alpha = 0.8) +
+ geom_text(
+ data = data,
+ mapping = aes(x = batch_nr, y = 0, label = features),
+ hjust = 0,
+ nudge_y = 0.05,
+ color = "white",
+ size = 5
+ ) +
+ coord_flip() +
+ xlab("Iteration") +
+ theme_minimal()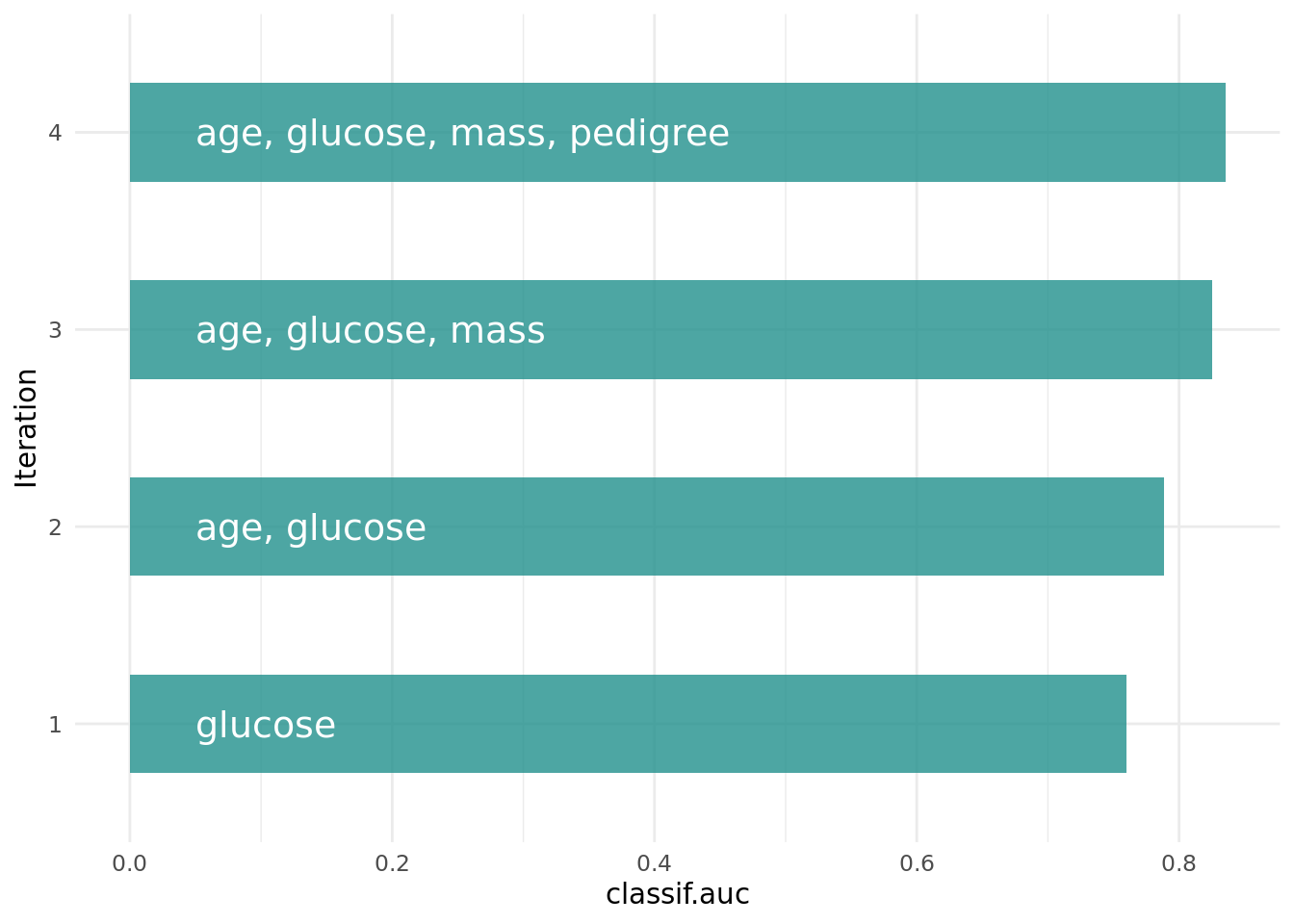 +
+The archive contains all evaluated feature sets. We can see that each feature has a corresponding shadow variable. We only show the variables age, glucose and insulin and their shadow variables here.
+as.data.table(instance$archive)[, .(age, glucose, insulin, permuted__age, permuted__glucose, permuted__insulin, classif.auc)] age glucose insulin permuted__age permuted__glucose permuted__insulin classif.auc
+ 1: TRUE FALSE FALSE FALSE FALSE FALSE 0.6437052
+ 2: FALSE TRUE FALSE FALSE FALSE FALSE 0.7598155
+ 3: FALSE FALSE TRUE FALSE FALSE FALSE 0.4900280
+ 4: FALSE FALSE FALSE FALSE FALSE FALSE 0.6424026
+ 5: FALSE FALSE FALSE FALSE FALSE FALSE 0.5690107
+---
+54: TRUE TRUE FALSE FALSE FALSE FALSE 0.8266713
+55: TRUE TRUE FALSE FALSE FALSE FALSE 0.8063568
+56: TRUE TRUE FALSE FALSE FALSE FALSE 0.8244232
+57: TRUE TRUE FALSE FALSE FALSE FALSE 0.8234605
+58: TRUE TRUE FALSE FALSE FALSE FALSE 0.8164784The learner we use to make predictions on new data is called the final model. The final model is trained with the optimal feature set on the full data set. We subset the task to the optimal feature set and train the learner.
+task$select(instance$result_feature_set)
+learner$train(task)The trained model can now be used to predict new, external data.
+The shadow variable search is a fast feature selection method that is easy to use. More information on the theoretical background can be found in Wu, Boos, and Stefanski (2007) and Thomas et al. (2017). If you want to know more about feature selection in general, we recommend having a look at our book.
+ + + +The predictive performance of modern machine learning algorithms is highly dependent on the choice of their hyperparameter configuration. Options for setting hyperparameters are tuning, manual selection by the user, and using the default configuration of the algorithm. The default configurations are chosen to work with a wide range of data sets but they usually do not achieve the best predictive performance. When tuning a learner in mlr3, we can run the default configuration as a baseline. Seeing how well it performs will tell us whether tuning pays off. If the optimized configurations perform worse, we could expand the search space or try a different optimization algorithm. Of course, it could also be that tuning on the given data set is simply not worth it.
+Probst, Boulesteix, and Bischl (2019) studied the tunability of machine learning algorithms. They found that the tunability of algorithms varies widely. Algorithms like glmnet and XGBoost are highly tunable, while algorithms like random forests work well with their default configuration. The highly tunable algorithms should thus beat their baselines more easily with optimized hyperparameters. In this article, we will tune the hyperparameters of a random forest and compare the performance of the default configuration with the optimized configurations.
+ +We tune the hyperparameters of the ranger learner on the spam data set. The search space is taken from Bischl et al. (2021).
library(mlr3verse)
+
+learner = lrn("classif.ranger",
+ mtry.ratio = to_tune(0, 1),
+ replace = to_tune(),
+ sample.fraction = to_tune(1e-1, 1),
+ num.trees = to_tune(1, 2000)
+)When creating the tuning instance, we set evaluate_default = TRUE to test the default hyperparameter configuration. The default configuration is evaluated in the first batch of the tuning run. The other batches use the specified tuning method. In this example, they are randomly drawn configurations.
instance = tune(
+ tuner = tnr("random_search", batch_size = 5),
+ task = tsk("spam"),
+ learner = learner,
+ resampling = rsmp ("holdout"),
+ measures = msr("classif.ce"),
+ term_evals = 51,
+ evaluate_default = TRUE
+)The default configuration is recorded in the first row of the archive. The other rows contain the results of the random search.
+as.data.table(instance$archive)[, .(batch_nr, mtry.ratio, replace, sample.fraction, num.trees, classif.ce)] batch_nr mtry.ratio replace sample.fraction num.trees classif.ce
+ 1: 1 0.122807018 TRUE 1.0000000 500 0.04954368
+ 2: 2 0.285388074 TRUE 0.1794772 204 0.06584094
+ 3: 2 0.097424099 FALSE 0.9475526 1441 0.04237288
+ 4: 2 0.008888587 FALSE 0.3216562 1868 0.08409387
+ 5: 2 0.335543330 TRUE 0.8122653 106 0.05345502
+---
+47: 11 0.788995735 FALSE 0.3692454 344 0.06258149
+48: 11 0.459305038 TRUE 0.3153485 1354 0.06258149
+49: 11 0.220334408 TRUE 0.9357554 817 0.05345502
+50: 11 0.868385877 TRUE 0.6743246 1040 0.06127771
+51: 11 0.015417312 FALSE 0.5627943 1836 0.08213820We plot the performances of the evaluated hyperparameter configurations. The blue line connects the best configuration of each batch. We see that the default configuration already performs well and the optimized configurations can not beat it.
+library(mlr3viz)
+
+autoplot(instance, type = "performance")
The time required to test the default configuration is negligible compared to the time required to run the hyperparameter optimization. It gives us a valuable indication of whether our tuning is properly configured. Running the default configuration as a baseline is a good practice that should be used in every tuning run.
+ + + +We continue working with the Hyperband optimization algorithm (Li et al. 2018). The previous post used the number of boosting iterations of an XGBoost model as the resource. However, Hyperband is not limited to machine learning algorithms that are trained iteratively. The resource can also be the number of features, the training time of a model, or the size of the training data set. In this post, we will tune a support vector machine and use the size of the training data set as the fidelity parameter. The time to train a support vector machine and the performance increases with the size of the data set. This makes the data set size a suitable fidelity parameter for Hyperband. This is the second part of the Hyperband series. The first part can be found here Hyperband Series - Iterative Training. If you don’t know much about Hyperband, check out the first post which explains the algorithm in detail. We assume that you are already familiar with tuning in the mlr3 ecosystem. If not, you should start with the book chapter on optimization or the Hyperparameter Optimization on the Palmer Penguins Data Set post. A little knowledge about mlr3pipelines is beneficial but not necessary to understand the example.
+ +In this post, we will optimize the hyperparameters of the support vector machine on the Sonar data set. We begin by constructing a classification machine by setting type to "C-classification".
library("mlr3verse")
+
+learner = lrn("classif.svm", id = "svm", type = "C-classification")The mlr3pipelines package features a PipeOp for subsampling.
po("subsample")PipeOp: <subsample> (not trained)
+values: <frac=0.6321, stratify=FALSE, replace=FALSE>
+Input channels <name [train type, predict type]>:
+ input [Task,Task]
+Output channels <name [train type, predict type]>:
+ output [Task,Task]The PipeOp controls the size of the training data set with the frac parameter. We connect the PipeOp with the learner and get a GraphLearner.
graph_learner = as_learner(
+ po("subsample") %>>%
+ learner
+)The graph learner subsamples and then fits a support vector machine on the data subset. The parameter set of the graph learner is a combination of the parameter sets of the PipeOp and learner.
as.data.table(graph_learner$param_set)[, .(id, lower, upper, levels)] id lower upper levels
+ 1: subsample.frac 0 Inf
+ 2: subsample.stratify NA NA TRUE,FALSE
+ 3: subsample.replace NA NA TRUE,FALSE
+ 4: svm.cachesize -Inf Inf
+ 5: svm.class.weights NA NA
+---
+15: svm.nu -Inf Inf
+16: svm.scale NA NA
+17: svm.shrinking NA NA TRUE,FALSE
+18: svm.tolerance 0 Inf
+19: svm.type NA NA C-classification,nu-classificationNext, we create the search space. We use TuneToken to mark which hyperparameters should be tuned. We have to prefix the hyperparameters with the id of the PipeOps. The subsample.frac is the fidelity parameter that must be tagged with "budget" in the search space. The data set size is increased from 3.7% to 100%. For the other hyperparameters, we took the search space for support vector machines from the Kuehn et al. (2018) article. This search space works for a wide range of data sets.
graph_learner$param_set$set_values(
+ subsample.frac = to_tune(p_dbl(3^-3, 1, tags = "budget")),
+ svm.kernel = to_tune(c("linear", "polynomial", "radial")),
+ svm.cost = to_tune(1e-4, 1e3, logscale = TRUE),
+ svm.gamma = to_tune(1e-4, 1e3, logscale = TRUE),
+ svm.tolerance = to_tune(1e-4, 2, logscale = TRUE),
+ svm.degree = to_tune(2, 5)
+)Support vector machines often crash or never finish the training with certain hyperparameter configurations. We set a timeout of 30 seconds and a fallback learner to handle these cases.
+graph_learner$encapsulate = c(train = "evaluate", predict = "evaluate")
+graph_learner$timeout = c(train = 30, predict = 30)
+graph_learner$fallback = lrn("classif.featureless")Let’s create the tuning instance. We use the "none" terminator because Hyperband controls the termination itself.
instance = ti(
+ task = tsk("sonar"),
+ learner = graph_learner,
+ resampling = rsmp("cv", folds = 3),
+ measures = msr("classif.ce"),
+ terminator = trm("none")
+)
+instance<TuningInstanceSingleCrit>
+* State: Not optimized
+* Objective: <ObjectiveTuning:subsample.svm_on_sonar>
+* Search Space:
+ id class lower upper nlevels
+1: subsample.frac ParamDbl 0.03703704 1.0000000 Inf
+2: svm.cost ParamDbl -9.21034037 6.9077553 Inf
+3: svm.degree ParamInt 2.00000000 5.0000000 4
+4: svm.gamma ParamDbl -9.21034037 6.9077553 Inf
+5: svm.kernel ParamFct NA NA 3
+6: svm.tolerance ParamDbl -9.21034037 0.6931472 Inf
+* Terminator: <TerminatorNone>We load the Hyperband tuner and set eta = 3.
library("mlr3hyperband")
+
+tuner = tnr("hyperband", eta = 3)Using eta = 3 and a lower bound of 3.7% for the data set size, results in the following schedule. Configurations with the same data set size are evaluated in parallel.
Now we are ready to start the tuning.
+tuner$optimize(instance)The best model is a support vector machine with a polynomial kernel.
+instance$result[, .(subsample.frac, svm.cost, svm.degree, svm.gamma, svm.kernel, svm.tolerance, classif.ce)] subsample.frac svm.cost svm.degree svm.gamma svm.kernel svm.tolerance classif.ce
+1: 1 1.871535 3 -2.60663 polynomial -4.573951 0.1491373The archive contains all evaluated configurations. We look at the 8 configurations that were evaluated on the complete data set. The configuration with the best classification error on the full data set was sampled in bracket 2. The classification error was estimated to be 26% on 33% of the data set and increased to 19% on the full data set (see green line in Figure 1).
+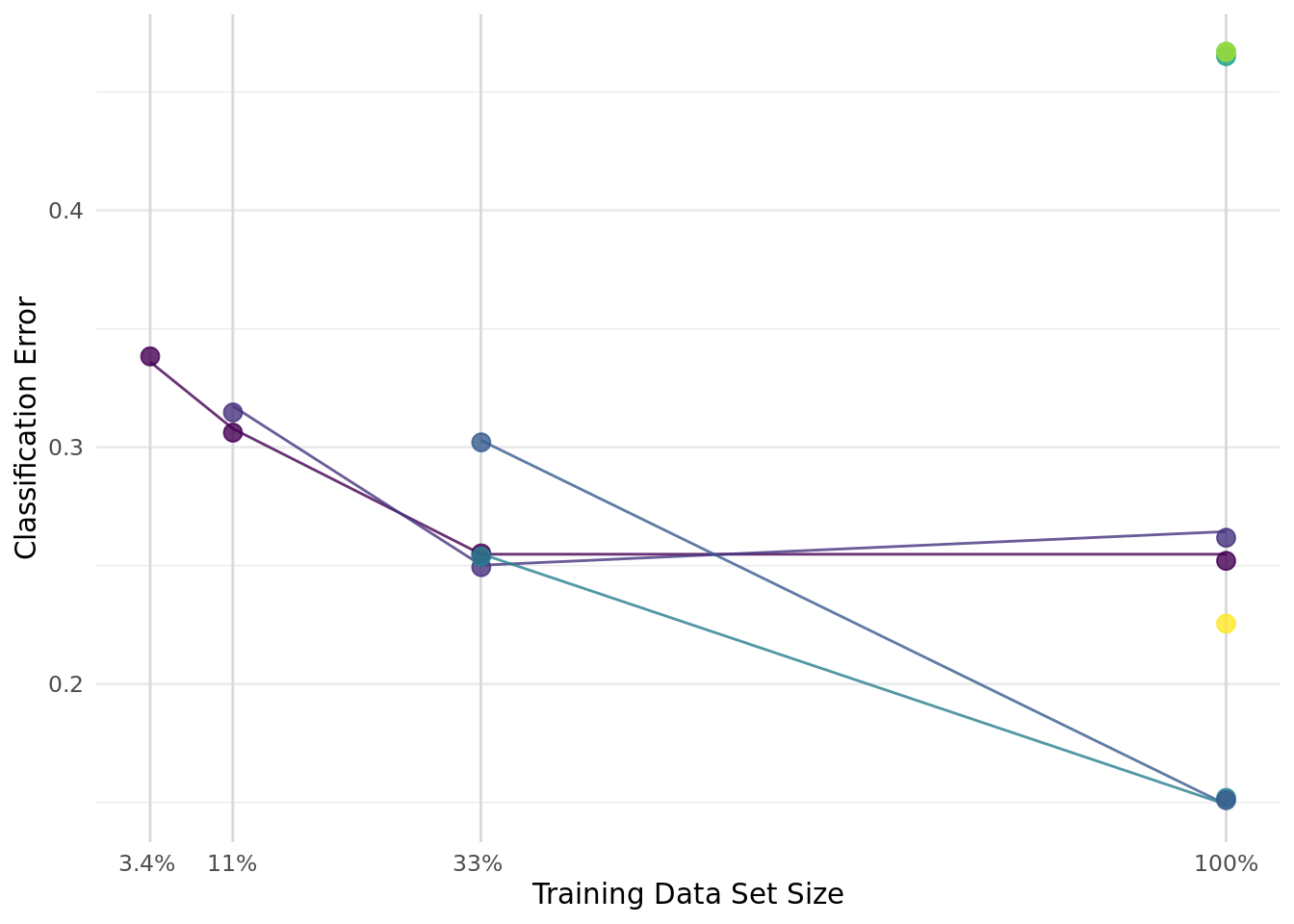 +
+Using the data set size as the budget parameter in Hyperband allows the tuning of machine learning models that are not trained iteratively. We have tried to keep the runtime of the example low. For your optimization, you should use cross-validation and run multiple iterations of Hyperband.
+Hotstarting a learner resumes the training from an already fitted model. An example would be to train an already fit XGBoost model for an additional 500 boosting iterations. In mlr3, we call this process Hotstarting, where a learner has access to a cache of already trained models which is called a mlr3::HoststartStack We distinguish between forward and backward hotstarting. We start this post with backward hotstarting and then talk about the less efficient forward hotstarting.
In this example, we optimize the hyperparameters of a random forest and use hotstarting to reduce the runtime. Hotstarting a random forest backwards is very simple. The model remains unchanged and only a subset of the trees is used for prediction i.e. a new model is not fitted. For example, a random forest is trained with 1000 trees and a specific hyperparameter configuration. If another random forest with 500 trees but with the same hyperparameter configuration has to be trained, the model with 1000 trees is copied and only 500 trees are used for prediction.
+We load the ranger learner and set the search space from the Bischl et al. (2021) article.
library(mlr3verse)
+
+learner = lrn("classif.ranger",
+ mtry.ratio = to_tune(0, 1),
+ replace = to_tune(),
+ sample.fraction = to_tune(1e-1, 1),
+ num.trees = to_tune(1, 2000)
+)We activate hotstarting with the allow_hotstart option. When running a grid search with hotstarting, the grid is sorted by the hot start parameter. This means the models with 2000 trees are trained first. The models with less than 2000 trees hot start on the 2000 trees models which allows the training to be completed immediately.
instance = tune(
+ tuner = tnr("grid_search", resolution = 5, batch_size = 5),
+ task = tsk("spam"),
+ learner = learner,
+ resampling = rsmp("holdout"),
+ measure = msr("classif.ce"),
+ allow_hotstart = TRUE
+)For comparison, we perform the same tuning without hotstarting.
+instance_2 = tune(
+ tuner = tnr("grid_search", resolution = 5, batch_size = 5),
+ task = tsk("spam"),
+ learner = learner,
+ resampling = rsmp("holdout"),
+ measure = msr("classif.ce"),
+ allow_hotstart = FALSE
+)We plot the time of completion of each batch (see Figure 1). Each batch includes 5 configurations. We can see that tuning with hotstarting is slower at first. As soon as all models are fitted with 2000 trees, the tuning runs much faster and overtakes the tuning without hotstarting.
+ +
+Forward hotstarting is currently only supported by XGBoost. However, we have observed that hotstarting only provides a speed advantage for very large datasets and models with more than 5000 boosting rounds. The reason is that copying the models from the main process to the workers is a major bottleneck. The parallelization package future copies the models sequentially to the workers. Consequently, it takes a long time until the last worker can even start. Moreover, copying itself consumes a lot of time, and copying the model back from the worker blocks the main process again. During the development process, we overestimated the speed benefits of hotstarting and underestimated the overhead of parallelization. We can therefore only advise against using forward hotstarting during tuning. It is much more efficient to use the internal early-stopping mechanism of XGBoost. This eliminates the need to copy models to the worker. See the gallery post on early stopping for an example. We might improve the efficiency of the hotstarting mechanism in the future, if there are convincing use cases.
+Nevertheless, forward hotstarting can be useful without parallelization. If you have an already trained model and want to add more boosting iteration to it. In this example, the learner_5000 is the already trained model. We create a new learner with the same hyperparameters but double the number of boosting iteration. To activate hotstarting, we create a HotstartStack and copy it to the $hotstart_stack slot of the new learner.
task = tsk("spam")
+
+learner_5000 = lrn("classif.xgboost", nrounds = 5000, eta = 0.1)
+learner_5000$train(task)
+
+learner_10000 = lrn("classif.xgboost", nrounds = 10000, eta = 0.1)
+learner_10000$hotstart_stack = HotstartStack$new(learner_5000)
+learner_10000$train(task)Training the initial model took 59.885 seconds.
+learner_5000$state$train_time[1] 59.885Adding 5000 boosting rounds took 46.837 seconds.
+learner_10000$state$train_time - learner_5000$state$train_time[1] 46.837Training the model from the beginning would have taken about two minutes. This means, without parallelization, we get the expected speed advantage.
+We have seen how mlr3 enables to reduce the training time, by building on a hotstart stack of already trained learners. One has to be careful, however, when using forward hotstarting during tuning because of the high parallelization overhead that arises from copying the models between the processes. If a model has an internal early stopping implementation, it should usually be relied upon instead of using the mlr3 hotstarting mechanism. However, manual forward hotstarting can be helpful in some situations when we do not want to train a large model from the beginning.
+ + + +Increasingly large data sets and search spaces make hyperparameter optimization a time-consuming task. Hyperband (Li et al. 2018) solves this by approximating the performance of a configuration on a simplified version of the problem such as a small subset of the training data, with just a few training epochs in a neural network, or with only a small number of iterations in a gradient-boosting model. After starting randomly sampled configurations, Hyperband iteratively allocates more resources to promising configurations and terminates low-performing ones. This type of optimization is called multi-fidelity optimization. The fidelity parameter is part of the search space and controls the tradeoff between the runtime and accuracy of the performance approximation. In this post, we will optimize XGBoost and use the number of boosting iterations as the fidelity parameter. This means Hyperband will allocate more boosting iterations to well-performing configurations. The number of boosting iterations increases the time to train a model and improves the performance until the model is overfitting to the training data. It is therefore a suitable fidelity parameter. We assume that you are already familiar with tuning in the mlr3 ecosystem. If not, you should start with the book chapter on optimization or the Hyperparameter Optimization on the Palmer Penguins Data Set post. This is the first part of the Hyperband series. The second part can be found here Hyperband Series - Data Set Subsampling.
+ +Hyperband is an advancement of the Successive Halving algorithm by Jamieson and Talwalkar (2016). Successive Halving is initialized with the number of starting configurations , the proportion of configurations discarded in each stage
, and the minimum
and maximum
budget of a single evaluation. The algorithm starts by sampling
random configurations and allocating the minimum budget
to them. The configurations are evaluated and
of the worst-performing configurations are discarded. The remaining configurations are promoted to the next stage and evaluated on a larger budget. This continues until one or more configurations are evaluated on the maximum budget
and the best performing configuration is selected. The number of stages is calculated so that each stage consumes approximately the same budget. This sometimes results in the minimum budget having to be slightly adjusted by the algorithm. Successive Halving has the disadvantage that is not clear whether we should choose a large
and try many configurations on a small budget or choose a small
and train more configurations on the full budget.
Hyperband solves this problem by running Successive Halving with different numbers of stating configurations. The algorithm is initialized with the same parameters as Successive Halving but without . Each run of Successive Halving is called a bracket and starts with a different budget
. A smaller starting budget means that more configurations can be tried out. The most explorative bracket allocated the minimum budget
. The next bracket increases the starting budget by a factor of
. In each bracket, the starting budget increases further until the last bracket
essentially performs a random search with the full budget
. The number of brackets
is calculated with
. Under the condition that
increases by
with each bracket,
sometimes has to be adjusted slightly in order not to use more than
resources in the last bracket. The number of configurations in the base stages is calculated so that each bracket uses approximately the same amount of budget. The following table shows a full run of the Hyperband algorithm. The bracket
is the most explorative bracket and
performance a random search on the full budget.
The Hyperband implementation in mlr3hyperband evaluates configurations with the same budget in parallel. This results in all brackets finishing at approximately the same time. The colors in Figure 1 indicate batches that are evaluated in parallel.
+In this practical example, we will optimize the hyperparameters of XGBoost on the Spam data set. We begin by loading the XGBoost learner..
library("mlr3verse")
+
+learner = lrn("classif.xgboost")The next thing we do is define the search space. The nrounds parameter controls the number of boosting iterations. We set a range from 16 to 128 boosting iterations. This is used as and
by the Hyperband algorithm. We need to tag the parameter with
"budget" to identify it as a fidelity parameter. For the other hyperparameters, we take the search space for XGBoost from the Bischl et al. (2021) article. This search space works for a wide range of data sets.
learner$param_set$set_values(
+ nrounds = to_tune(p_int(16, 128, tags = "budget")),
+ eta = to_tune(1e-4, 1, logscale = TRUE),
+ max_depth = to_tune(1, 20),
+ colsample_bytree = to_tune(1e-1, 1),
+ colsample_bylevel = to_tune(1e-1, 1),
+ lambda = to_tune(1e-3, 1e3, logscale = TRUE),
+ alpha = to_tune(1e-3, 1e3, logscale = TRUE),
+ subsample = to_tune(1e-1, 1)
+)We construct the tuning instance. We use the "none" terminator because Hyperband terminates itself when all brackets are evaluated.
instance = ti(
+ task = tsk("spam"),
+ learner = learner,
+ resampling = rsmp("holdout"),
+ measures = msr("classif.ce"),
+ terminator = trm("none")
+)
+instance<TuningInstanceSingleCrit>
+* State: Not optimized
+* Objective: <ObjectiveTuning:classif.xgboost_on_spam>
+* Search Space:
+ id class lower upper nlevels
+ <char> <char> <num> <num> <num>
+1: nrounds ParamInt 16.000000 128.000000 113
+2: eta ParamDbl -9.210340 0.000000 Inf
+3: max_depth ParamInt 1.000000 20.000000 20
+4: colsample_bytree ParamDbl 0.100000 1.000000 Inf
+5: colsample_bylevel ParamDbl 0.100000 1.000000 Inf
+6: lambda ParamDbl -6.907755 6.907755 Inf
+7: alpha ParamDbl -6.907755 6.907755 Inf
+8: subsample ParamDbl 0.100000 1.000000 Inf
+* Terminator: <TerminatorNone>We load the Hyperband tuner and set eta = 2. Hyperband can start from the beginning when the last bracket is evaluated. We control the number of Hyperband runs with the repetition argument. The setting repetition = Inf is useful when a terminator should stop the optimization.
library("mlr3hyperband")
+
+tuner = tnr("hyperband", eta = 2, repetitions = 1)The Hyperband implementation in mlr3hyperband evaluates configurations with the same budget in parallel. This results in all brackets finishing at approximately the same time. You can think of it as going diagonally through Figure 1. Using eta = 2 and a range from 16 to 128 boosting iterations results in the following schedule.
Now we are ready to start the tuning.
+tuner$optimize(instance)The result of a run is the configuration with the best performance. This does not necessarily have to be a configuration evaluated with the highest budget since we can overfit the data with too many boosting iterations.
+instance$result[, .(nrounds, eta, max_depth, colsample_bytree, colsample_bylevel, lambda, alpha, subsample)] nrounds eta max_depth colsample_bytree colsample_bylevel lambda alpha subsample
+ <num> <num> <int> <num> <num> <num> <num> <num>
+1: 128 -1.985313 8 0.4828647 0.3001509 -3.816118 -4.998944 0.4464856The archive of a Hyperband run has the additional columns "bracket" and "stage".
as.data.table(instance$archive)[, .(bracket, stage, classif.ce, eta, max_depth, colsample_bytree)] bracket stage classif.ce eta max_depth colsample_bytree
+ <int> <num> <num> <num> <int> <num>
+ 1: 3 0 0.09061278 -3.6110452 20 0.2671318
+ 2: 3 0 0.06388527 -1.9853130 8 0.4828647
+ 3: 3 0 0.11147327 -2.8428844 3 0.3152709
+ 4: 3 0 0.07301173 -3.9228523 19 0.5369803
+ 5: 3 0 0.07953064 -3.6486183 17 0.2039727
+---
+31: 0 0 0.06258149 -1.8578637 9 0.7343156
+32: 3 3 0.03976532 -1.9853130 8 0.4828647
+33: 2 2 0.03976532 -0.9898459 4 0.2534038
+34: 1 1 0.05345502 -2.5345314 13 0.5260946
+35: 1 1 0.06714472 -8.5597326 16 0.9890136The handling of Hyperband in mlr3tuning is very similar to that of other tuners. We only have to select an additional fidelity parameter and tag it with "budget". We have tried to keep the runtime of the example low. For your optimization, you should use cross-validation and increase the maximum number of boosting rounds. The Bischl et al. (2021) search space suggests 5000 boosting rounds. Check out our next post on Hyperband which uses the size of the training data set as the fidelity parameter.
We showcase the visualization functions of the mlr3 ecosystem. The mlr3viz package creates a plot for almost all mlr3 objects. This post displays all available plots with their reproducible code. We start with plots of the base mlr3 objects. This includes boxplots of tasks, dendrograms of cluster learners and ROC curves of predictions. After that, we tune a classification tree and visualize the results. Finally, we show visualizations for filters.
+This article will be updated whenever a new plot is available in mlr3viz.
The mlr3viz package defines autoplot() functions to draw plots with ggplot2. Often there is more than one type of plot for an object. You can change the plot with the type argument. The help pages list all possible choices. The easiest way to access the help pages is via the pkgdown website. The plots use the viridis color pallet and the appearance is controlled with the theme argument. By default, the minimal theme is applied.
We begin with plots of the classification task Palmer Penguins. We plot the class frequency of the target variable.
library(mlr3verse)
+library(mlr3viz)
+
+task = tsk("penguins")
+task$select(c("body_mass", "bill_length"))
+
+autoplot(task, type = "target")
The "duo" plot shows the distribution of multiple features.
autoplot(task, type = "duo")
The "pairs" plot shows the pairwise comparison of multiple features. The classes of the target variable are shown in different colors.
autoplot(task, type = "pairs")
Next, we plot the regression task mtcars. We create a boxplot of the target variable.
task = tsk("mtcars")
+task$select(c("am", "carb"))
+
+autoplot(task, type = "target")
The "pairs" plot shows the pairwise comparison of mutiple features and the target variable.
autoplot(task, type = "pairs")
Finally, we plot the cluster task US Arrests. The "pairs" plot shows the pairwise comparison of mutiple features.
library(mlr3cluster)
+
+task = mlr_tasks$get("usarrests")
+
+autoplot(task, type = "pairs")
The "prediction" plot shows the decision boundary of a classification learner and the true class labels as points.
task = tsk("pima")$select(c("age", "pedigree"))
+learner = lrn("classif.rpart")
+learner$train(task)
+
+autoplot(learner, type = "prediction", task)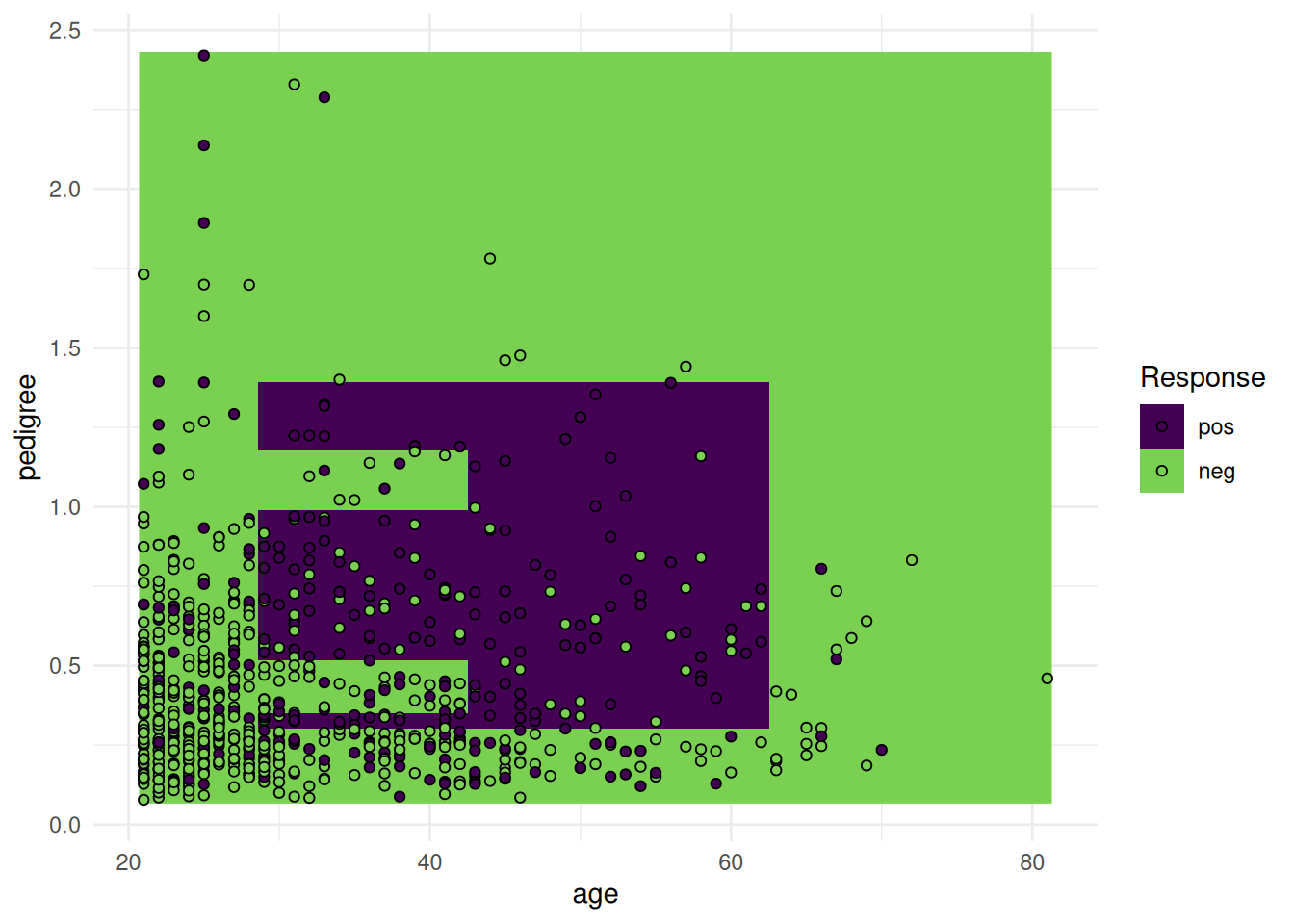
Using probabilities.
+task = tsk("pima")$select(c("age", "pedigree"))
+learner = lrn("classif.rpart", predict_type = "prob")
+learner$train(task)
+
+autoplot(learner, type = "prediction", task)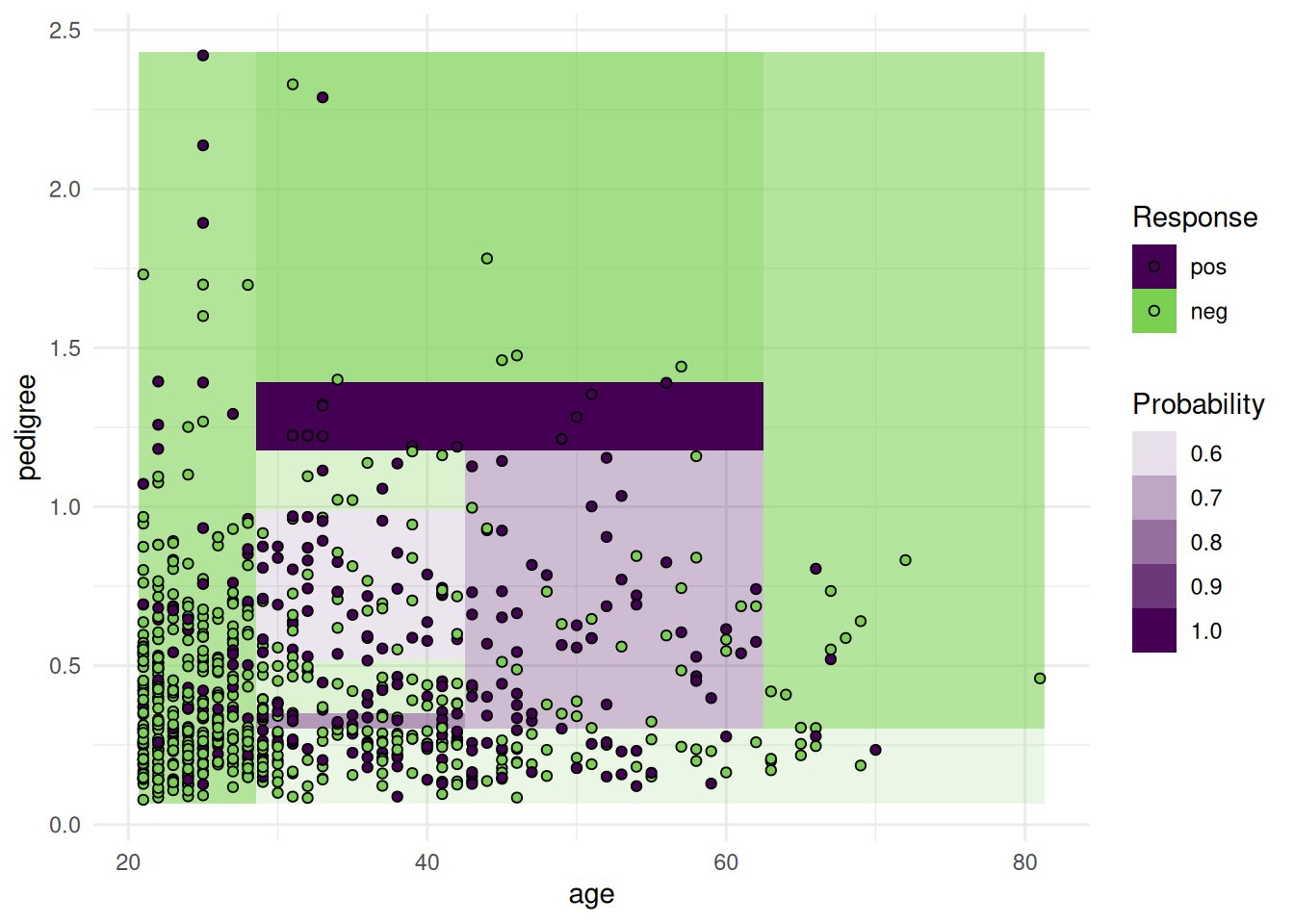
The "prediction" plot of a regression learner illustrates the decision boundary and the true response as points.
task = tsk("boston_housing")$select("age")
+learner = lrn("regr.rpart")
+learner$train(task)
+
+autoplot(learner, type = "prediction", task)
When using two features, the response surface is plotted in the background.
+task = tsk("boston_housing")$select(c("age", "rm"))
+learner = lrn("regr.rpart")
+learner$train(task)
+
+autoplot(learner, type = "prediction", task)
The classification and regression GLMNet learner is equipped with a plot function.
library(mlr3data)
+
+task = tsk("ilpd")
+task$select(setdiff(task$feature_names, "gender"))
+learner = lrn("classif.glmnet")
+learner$train(task)
+
+autoplot(learner, type = "ggfortify")
task = tsk("mtcars")
+learner = lrn("regr.glmnet")
+learner$train(task)
+
+autoplot(learner, type = "ggfortify")
We plot a classification tree of the rpart package. We have to fit the learner with keep_model = TRUE to keep the model object.
task = tsk("penguins")
+learner = lrn("classif.rpart", keep_model = TRUE)
+learner$train(task)
+
+autoplot(learner, type = "ggparty")
We can also plot regression trees.
+task = tsk("mtcars")
+learner = lrn("regr.rpart", keep_model = TRUE)
+learner$train(task)
+
+autoplot(learner, type = "ggparty")
The "dend" plot shows the result of the hierarchical clustering of the data.
library(mlr3cluster)
+
+task = tsk("usarrests")
+learner = lrn("clust.hclust")
+learner$train(task)
+
+autoplot(learner, type = "dend", task = task)
The "scree" type plots the number of clusters and the height.
autoplot(learner, type = "scree")
We plot the predictions of a classification learner. The "stacked" plot shows the predicted and true class labels.
task = tsk("spam")
+learner = lrn("classif.rpart", predict_type = "prob")
+pred = learner$train(task)$predict(task)
+
+autoplot(pred, type = "stacked")
The ROC curve plots the true positive rate against the false positive rate at different thresholds.
+autoplot(pred, type = "roc")
The precision-recall curve plots the precision against the recall at different thresholds.
+autoplot(pred, type = "prc")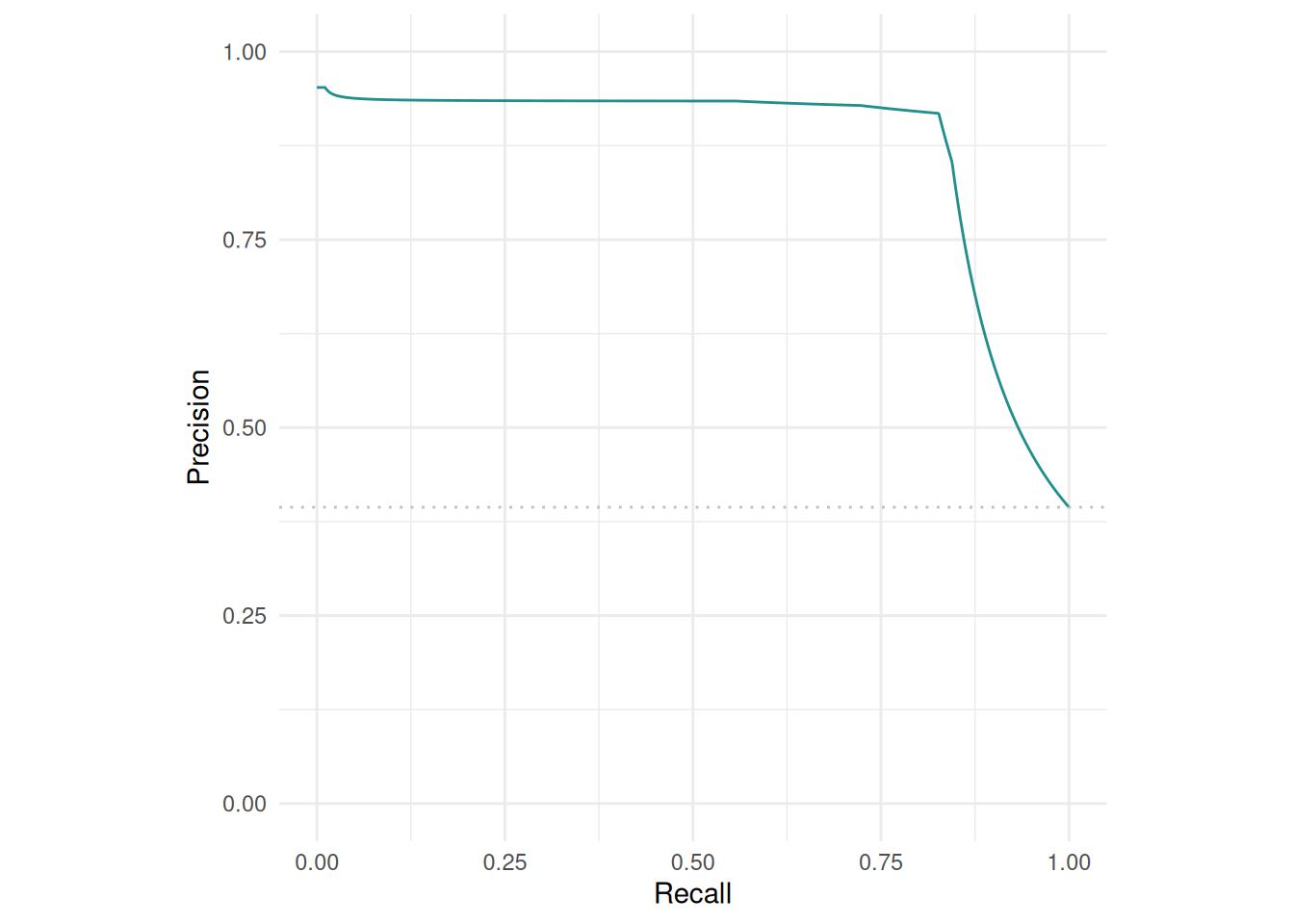
The "threshold" plot varies the threshold of a binary classification and plots against the resulting performance.
autoplot(pred, type = "threshold")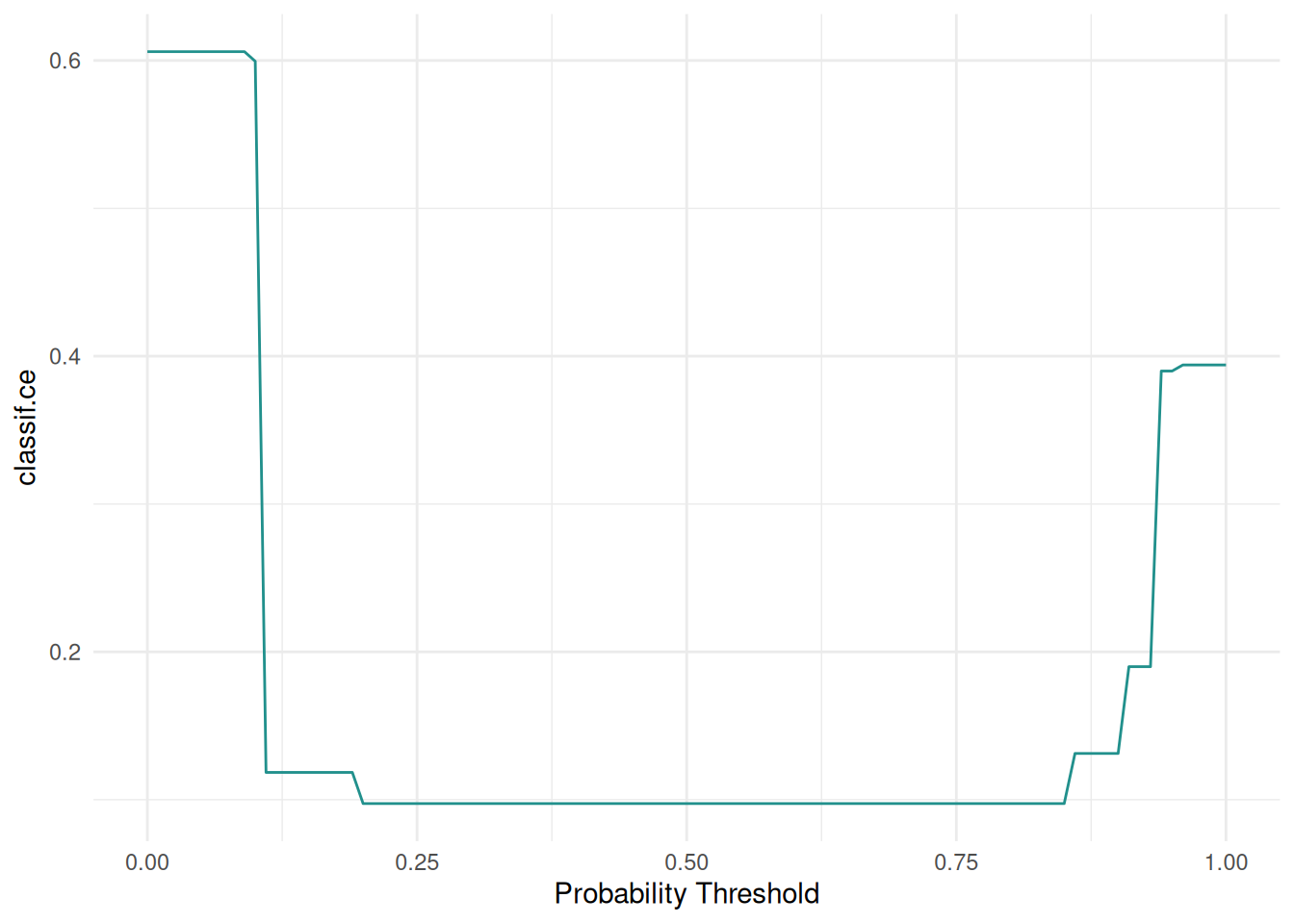
The predictions of a regression learner are often presented as a scatterplot of truth and predicted response.
+task = tsk("boston_housing")
+learner = lrn("regr.rpart")
+pred = learner$train(task)$predict(task)
+
+autoplot(pred, type = "xy")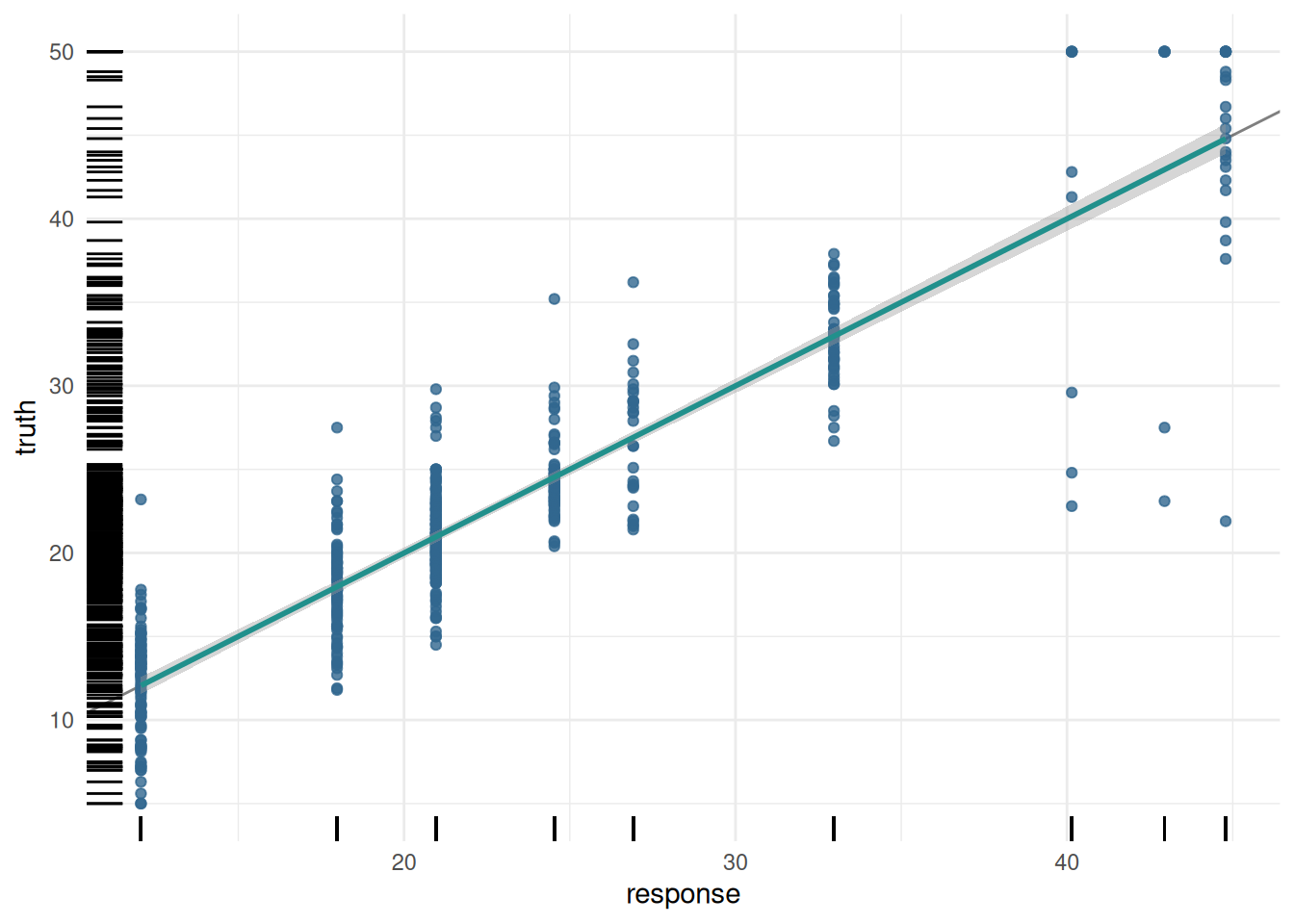
Additionally, we plot the response with the residuals.
+autoplot(pred, type = "residual")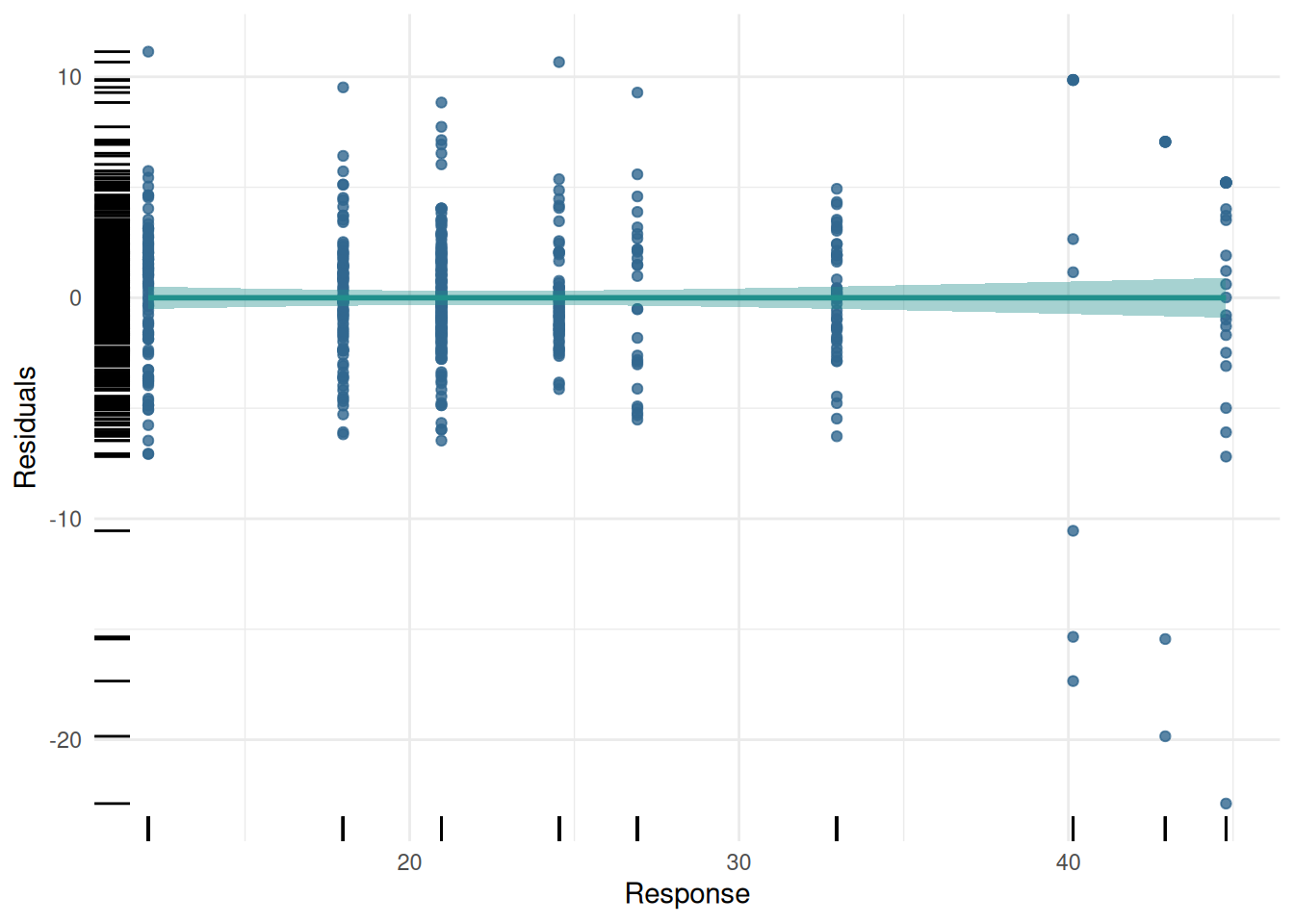
We can also plot the distribution of the residuals.
+autoplot(pred, type = "histogram")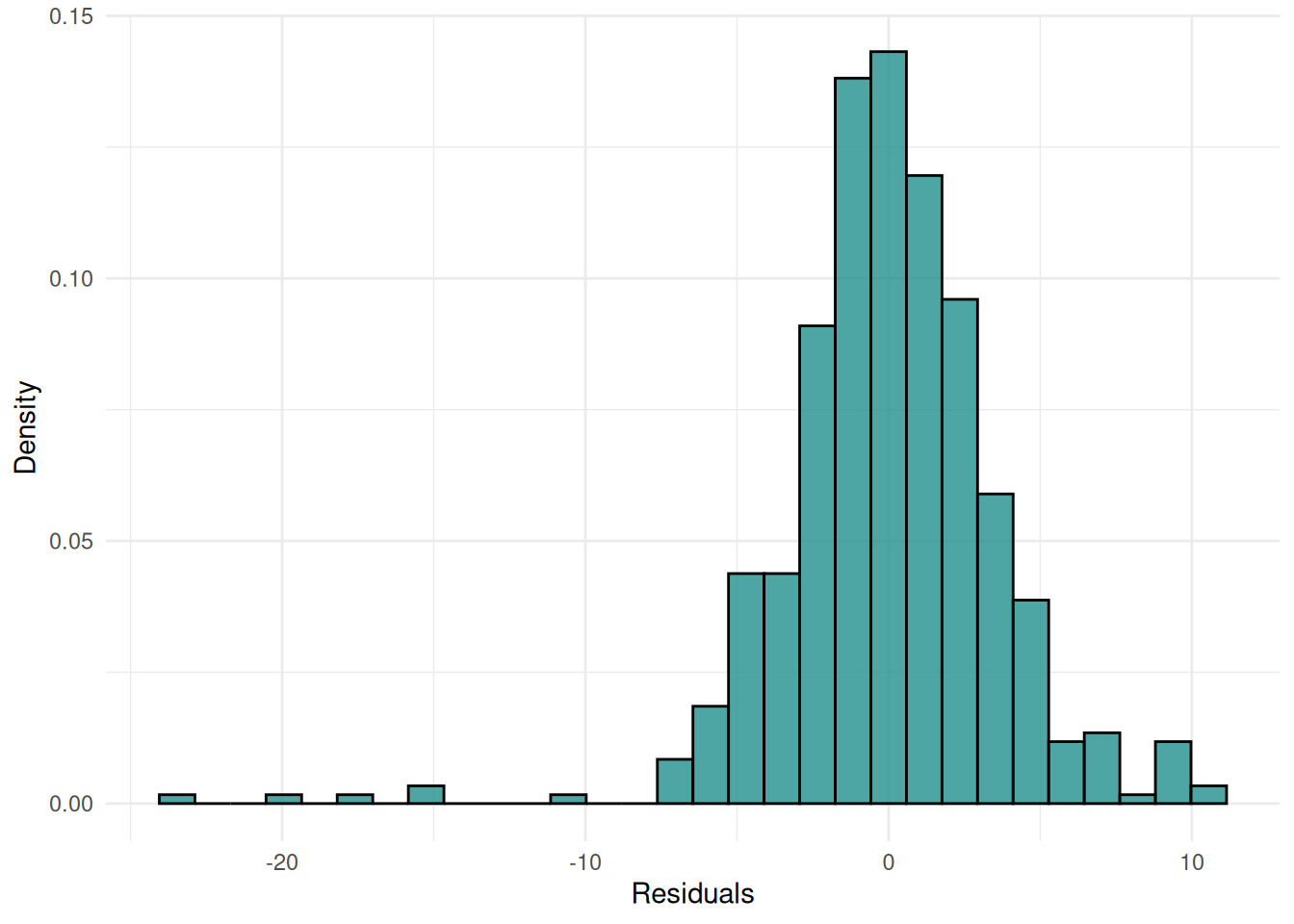
The predictions of a cluster learner are often presented as a scatterplot of the data points colored by the cluster.
+library(mlr3cluster)
+
+task = tsk("usarrests")
+learner = lrn("clust.kmeans", centers = 3)
+pred = learner$train(task)$predict(task)
+
+autoplot(pred, task, type = "scatter")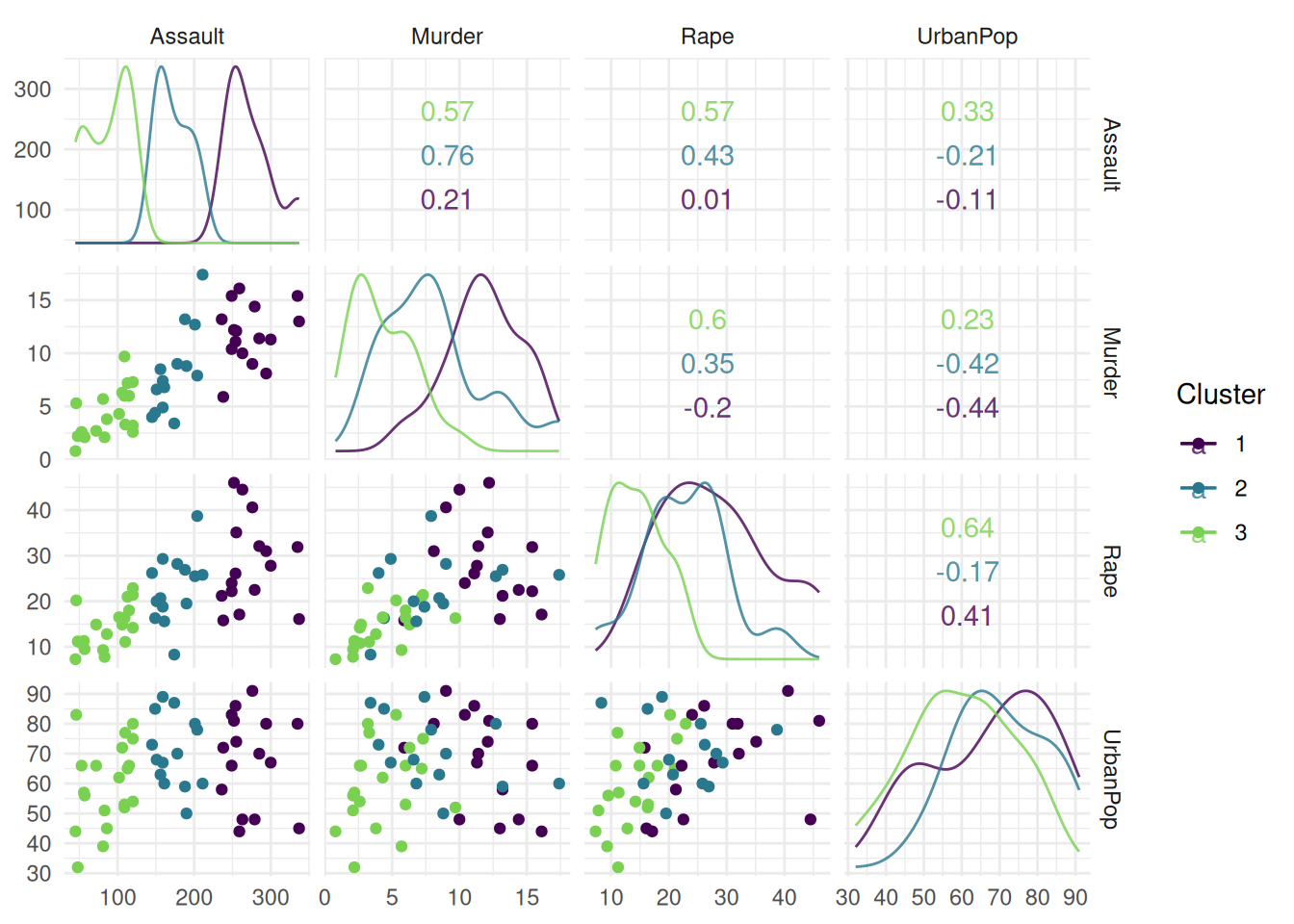
The "sil" plot shows the silhouette width of the clusters. The dashed line is the mean silhouette width.
autoplot(pred, task, type = "sil")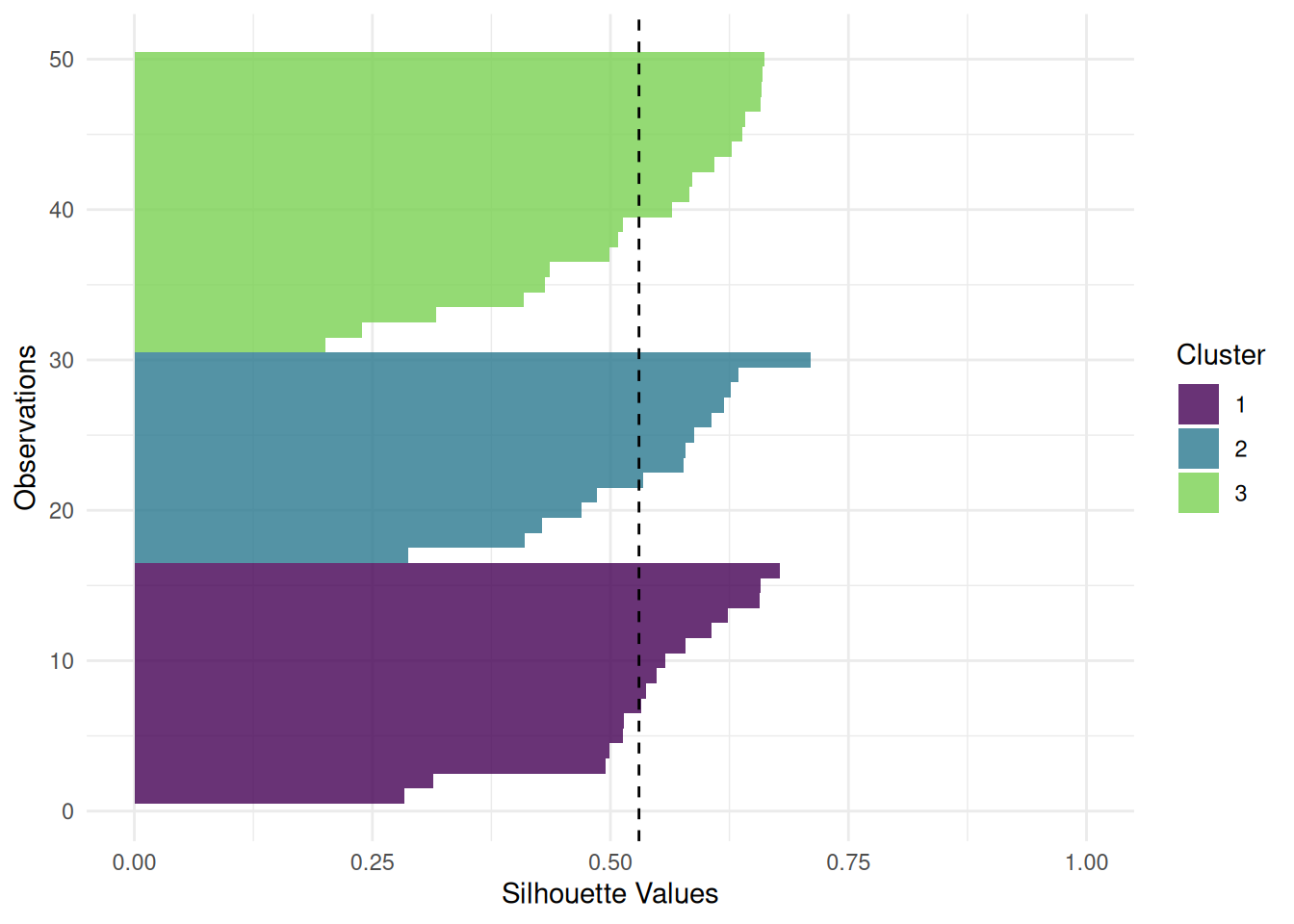
The "pca" plot shows the first two principal components of the data colored by the cluster.
autoplot(pred, task, type = "pca")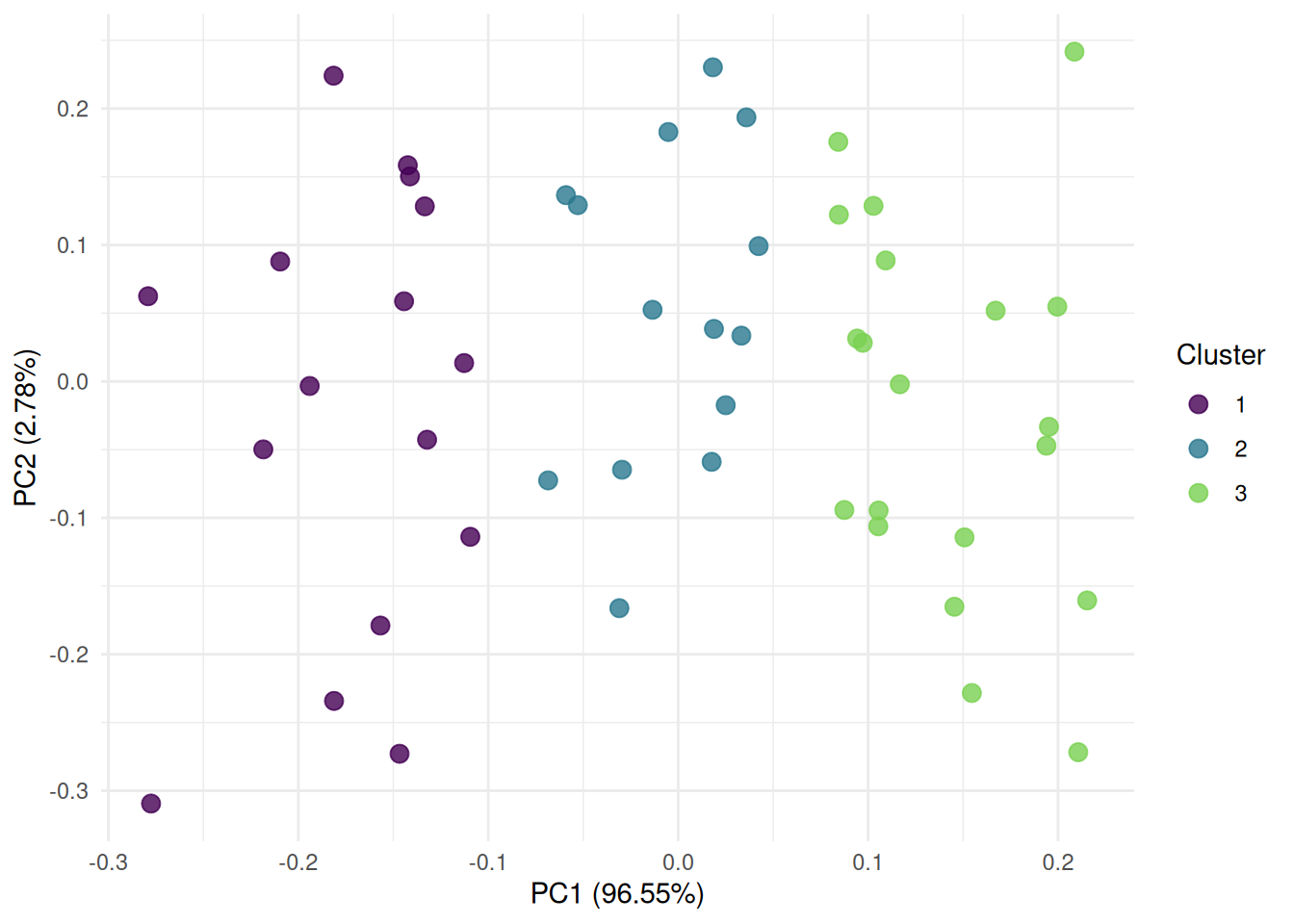
The "boxplot" shows the distribution of the performance measures.
task = tsk("sonar")
+learner = lrn("classif.rpart", predict_type = "prob")
+resampling = rsmp("cv")
+rr = resample(task, learner, resampling)
+
+autoplot(rr, type = "boxplot")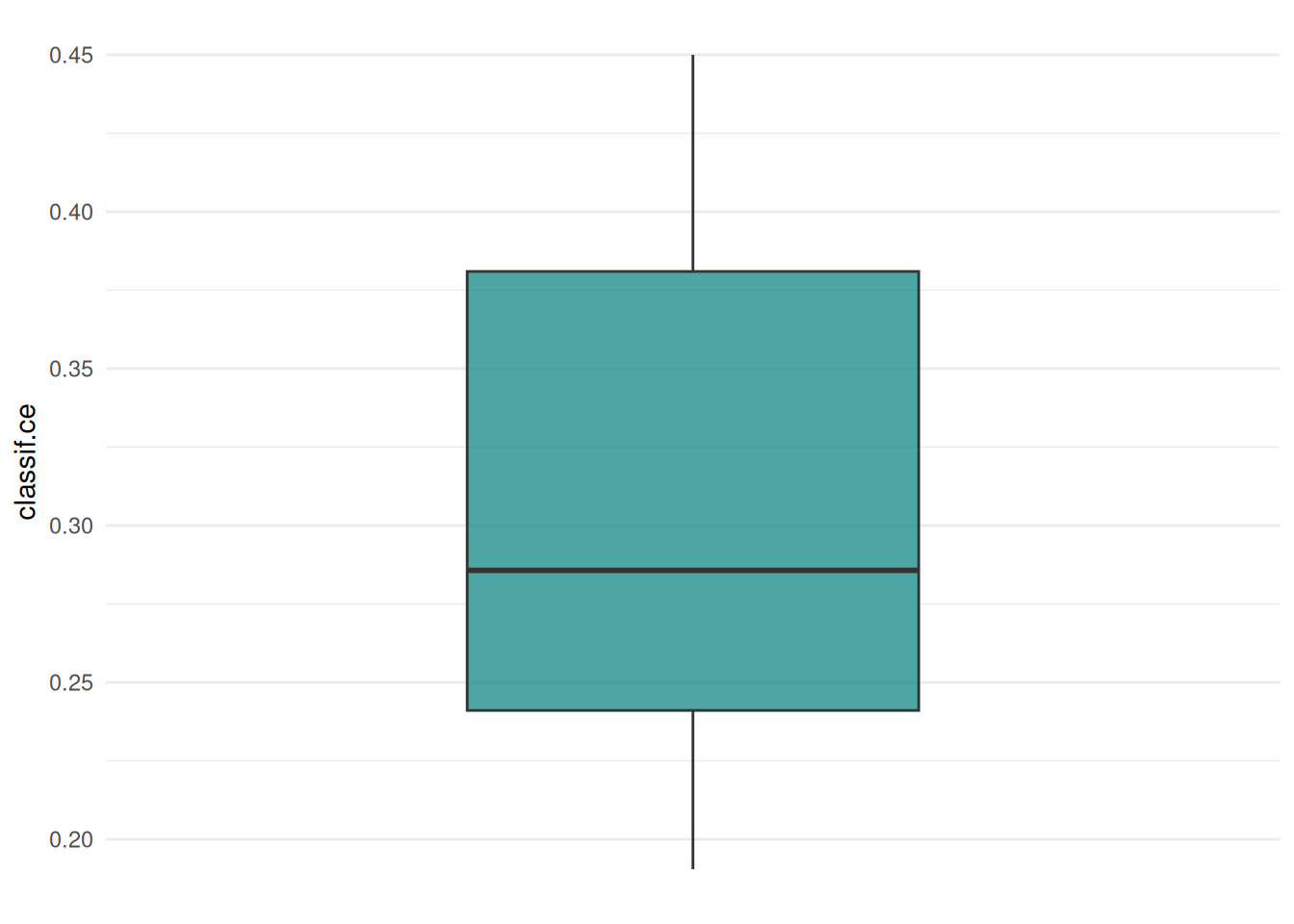
We can also plot the distribution of the performance measures as a “histogram”.
autoplot(rr, type = "histogram")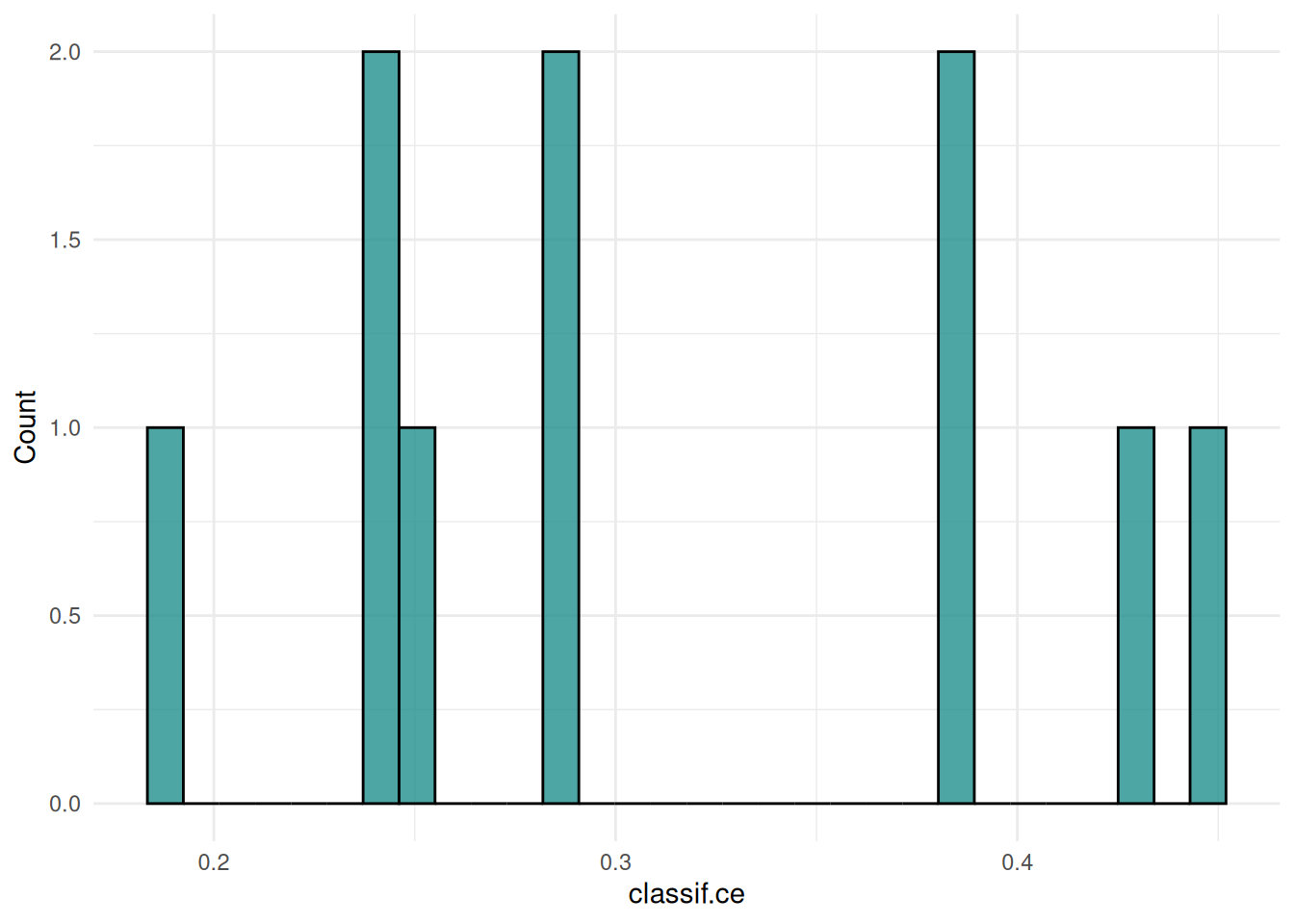
The ROC curve plots the true positive rate against the false positive rate at different thresholds.
+autoplot(rr, type = "roc")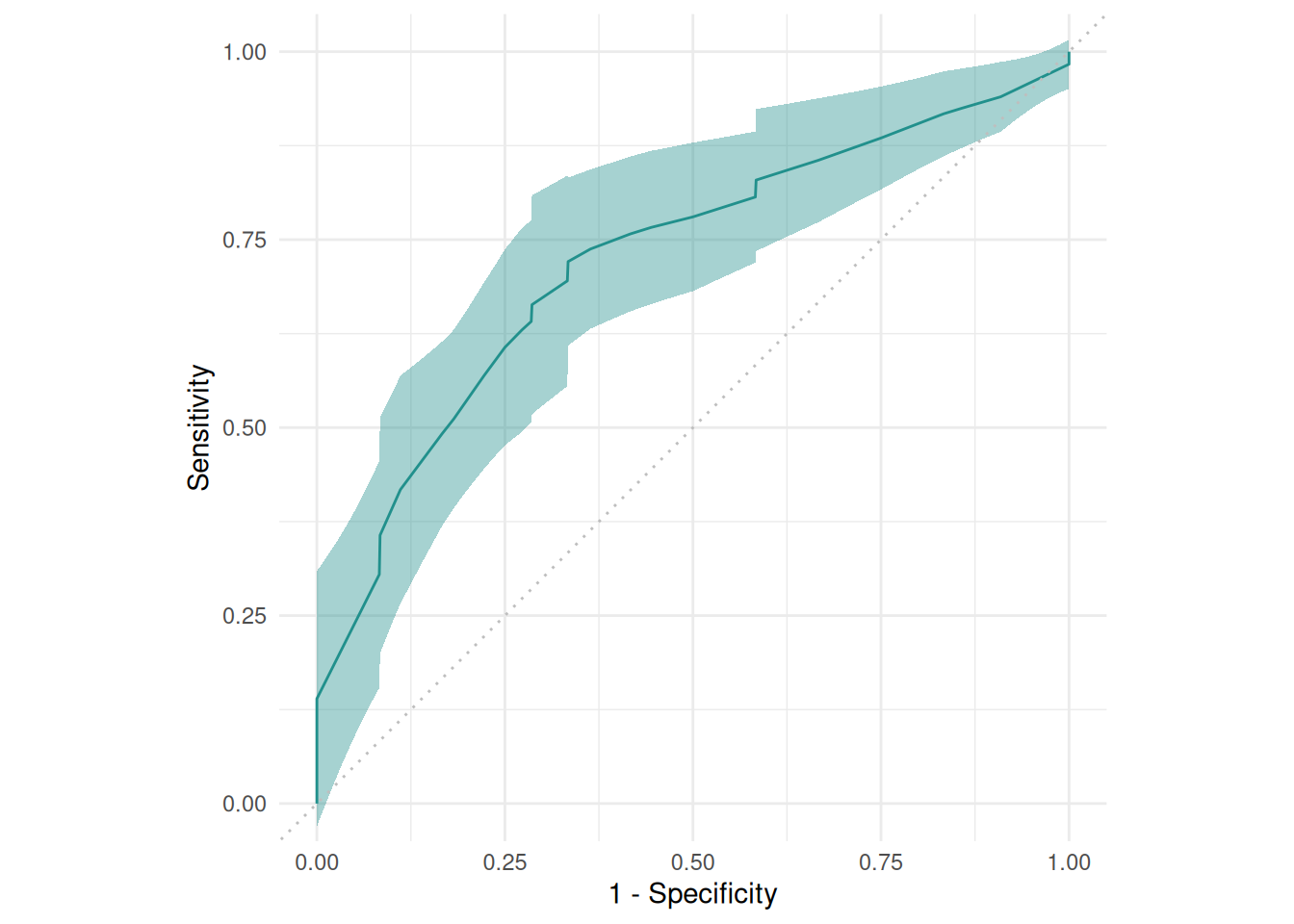
The precision-recall curve plots the precision against the recall at different thresholds.
+autoplot(rr, type = "prc")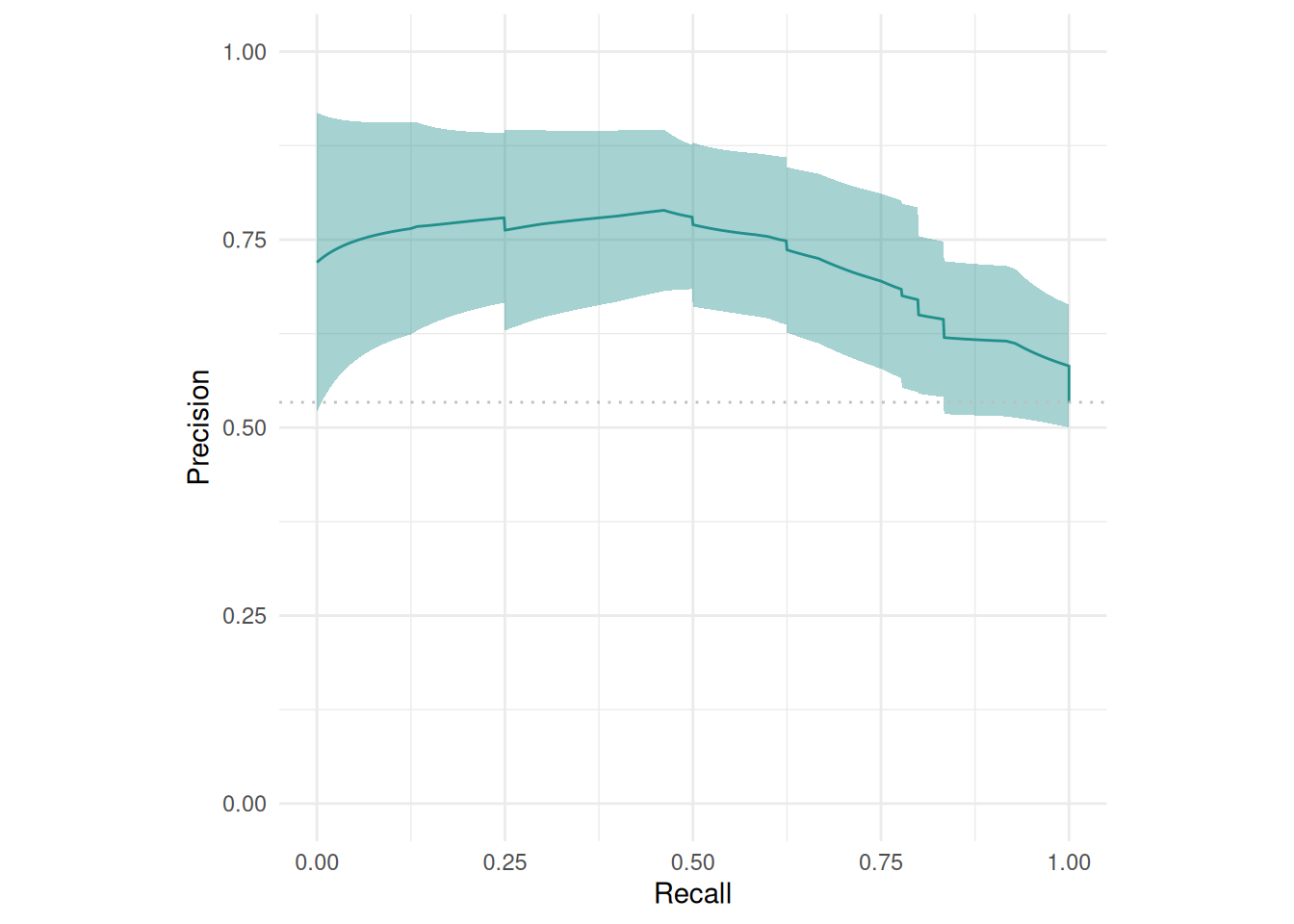
The "prediction" plot shows two features and the predicted class in the background. Points mark the observations of the test set and the color presents the truth.
task = tsk("pima")
+task$filter(seq(100))
+task$select(c("age", "glucose"))
+learner = lrn("classif.rpart")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")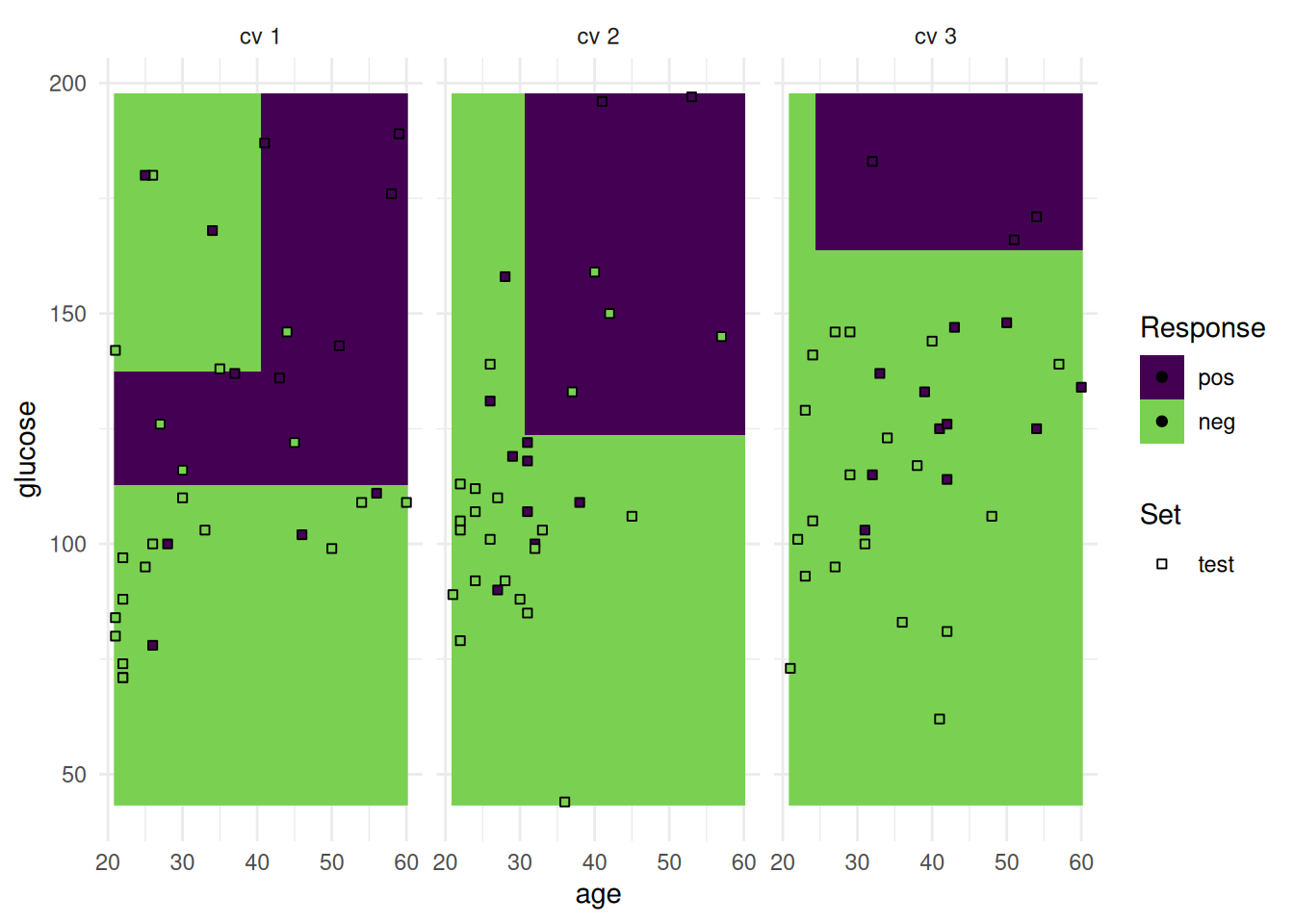
Alternatively, we can plot class probabilities.
+task = tsk("pima")
+task$filter(seq(100))
+task$select(c("age", "glucose"))
+learner = lrn("classif.rpart", predict_type = "prob")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")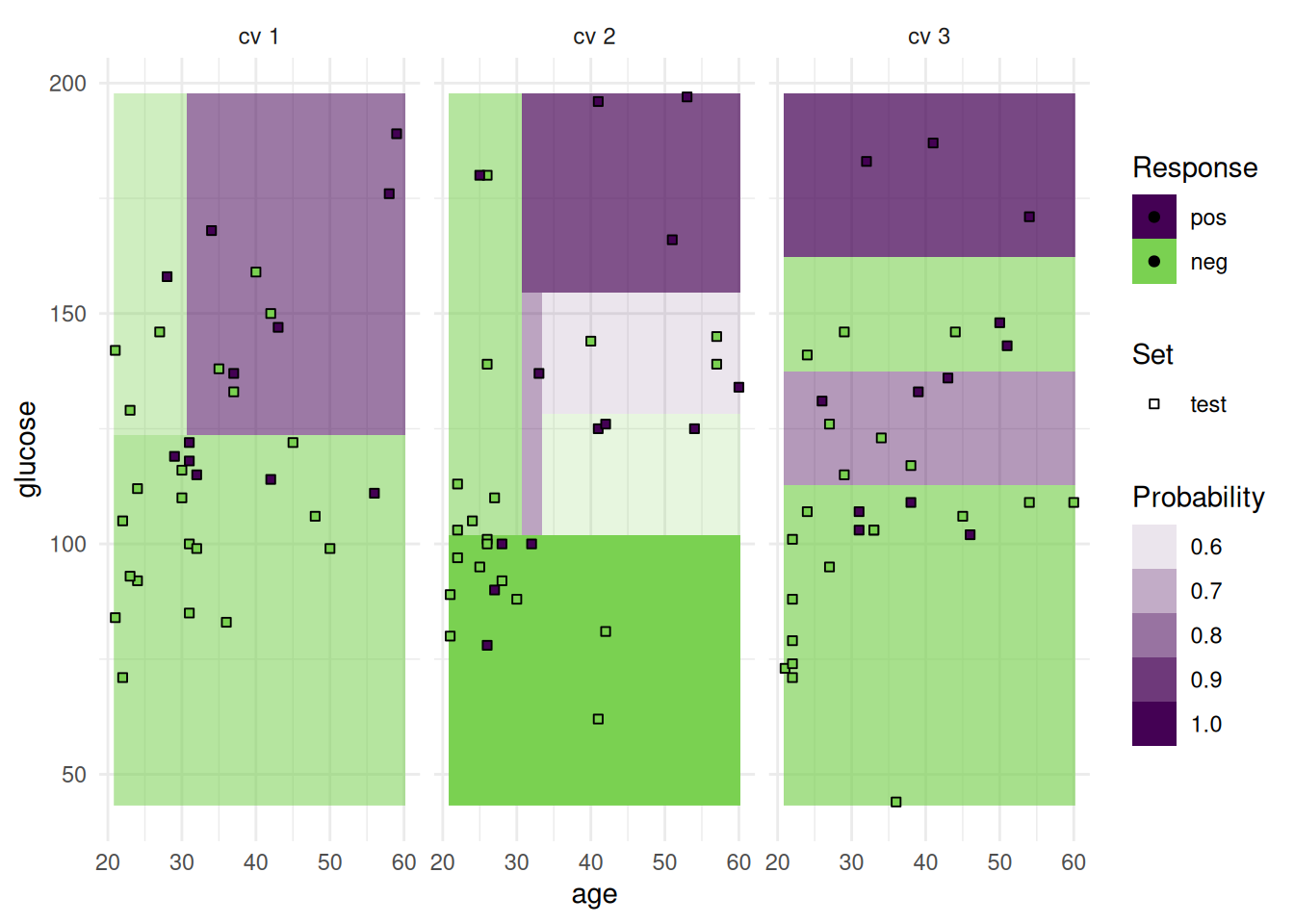
In addition to the test set, we can also plot the train set.
+task = tsk("pima")
+task$filter(seq(100))
+task$select(c("age", "glucose"))
+learner = lrn("classif.rpart", predict_type = "prob", predict_sets = c("train", "test"))
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction", predict_sets = c("train", "test"))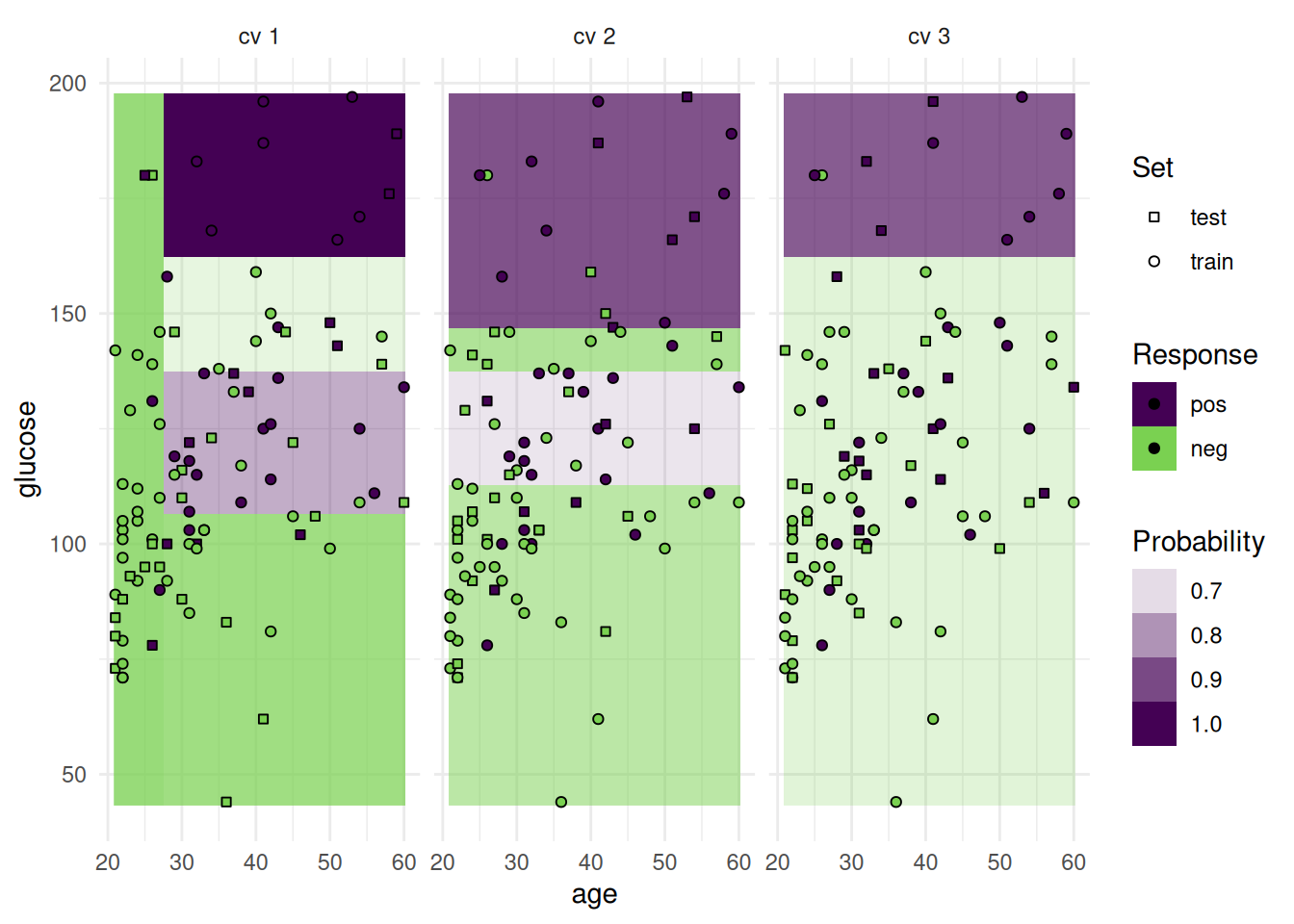
The "prediction" plot can also show categorical features.
task = tsk("german_credit")
+task$filter(seq(100))
+task$select(c("housing", "employment_duration"))
+learner = lrn("classif.rpart")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")
The “prediction” plot shows one feature and the response. Points mark the observations of the test set.
task = tsk("boston_housing")
+task$select("age")
+task$filter(seq(100))
+learner = lrn("regr.rpart")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")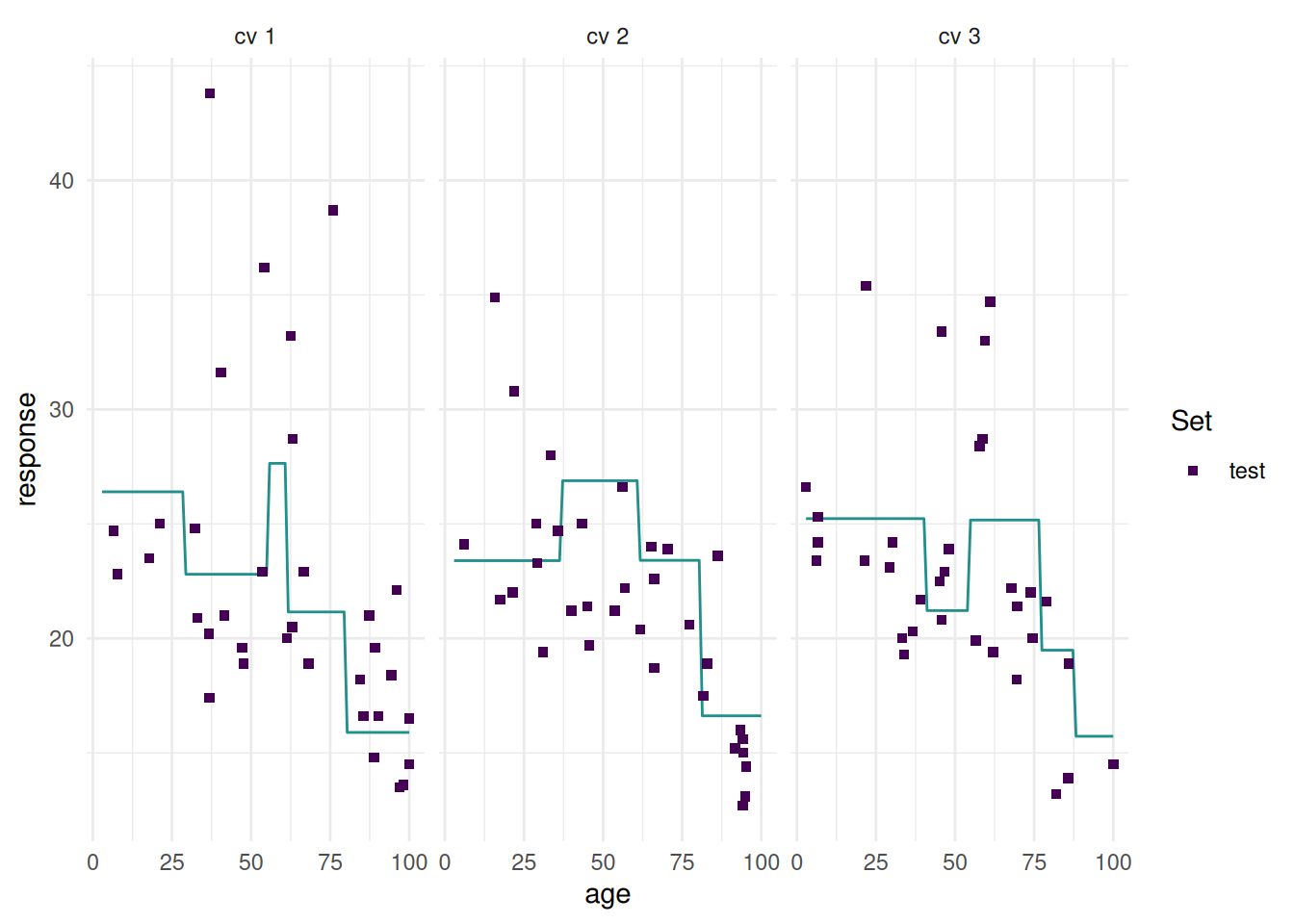
Additionally, we can add confidence bounds.
+task = tsk("boston_housing")
+task$select("age")
+task$filter(seq(100))
+learner = lrn("regr.lm", predict_type = "se")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")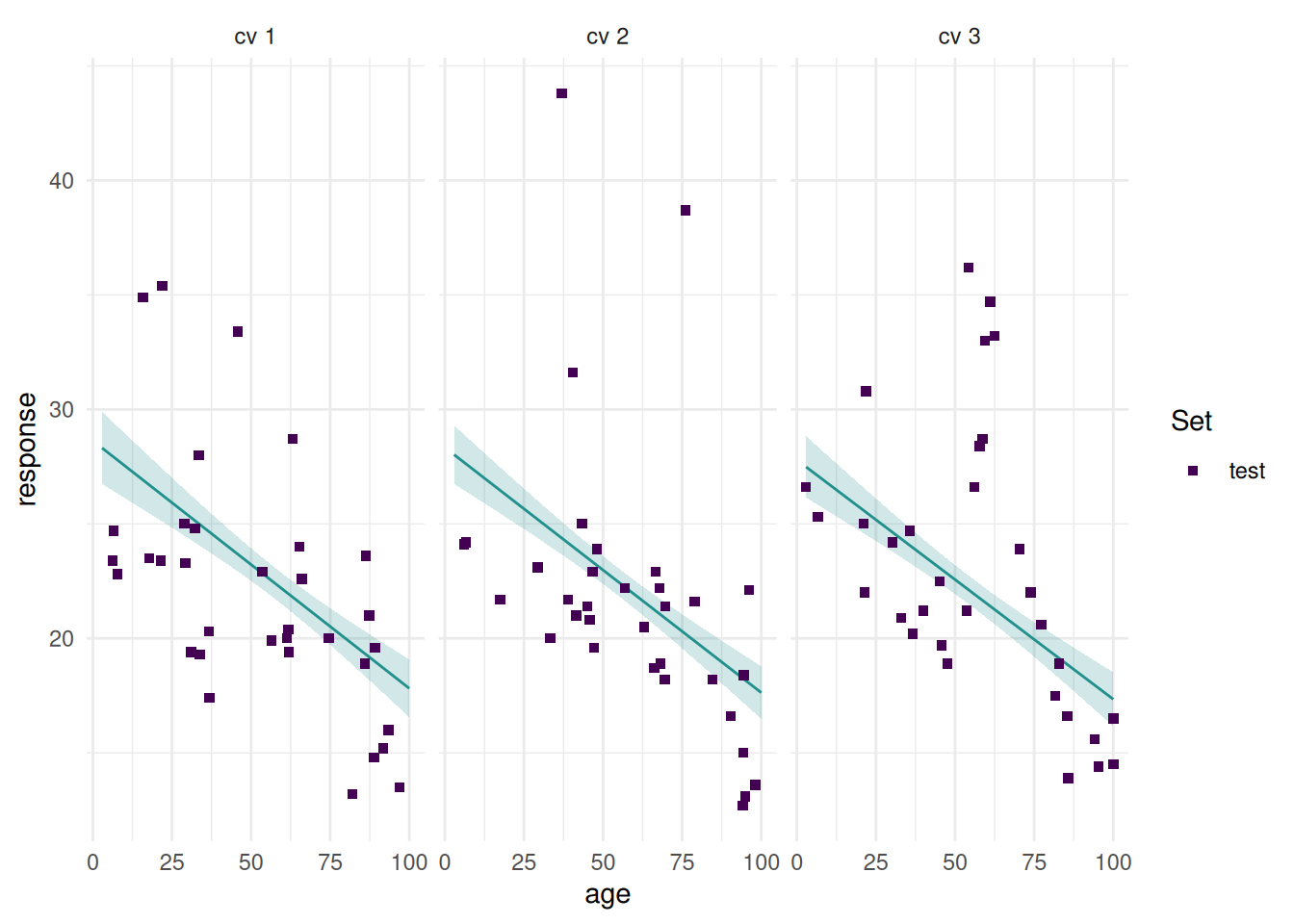
And add the train set.
+task = tsk("boston_housing")
+task$select("age")
+task$filter(seq(100))
+learner = lrn("regr.lm", predict_type = "se", predict_sets = c("train", "test"))
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction", predict_sets = c("train", "test"))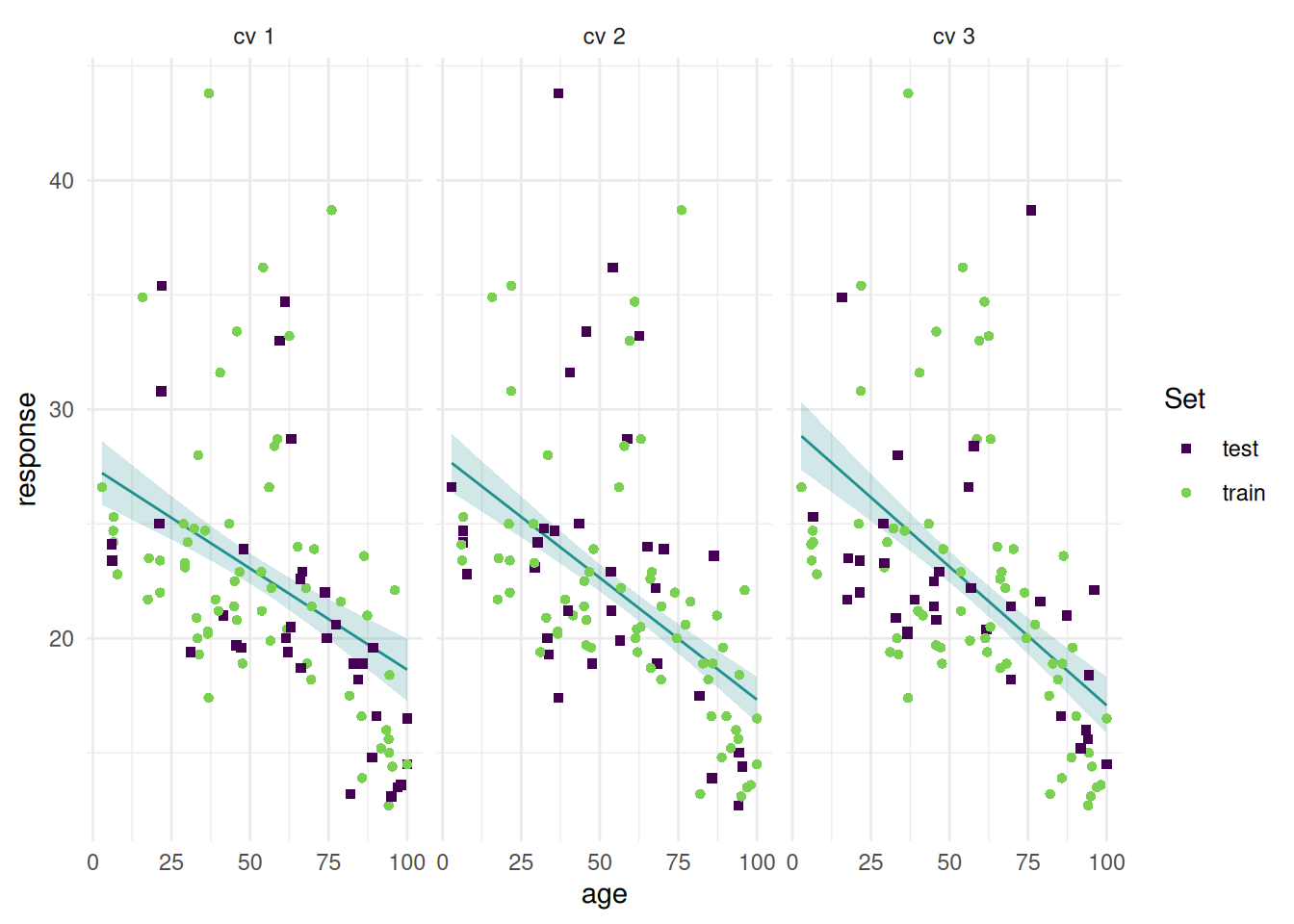
We can also add the prediction surface to the background.
+task = tsk("boston_housing")
+task$select(c("age", "rm"))
+task$filter(seq(100))
+learner = lrn("regr.rpart")
+resampling = rsmp("cv", folds = 3)
+rr = resample(task, learner, resampling, store_models = TRUE)
+
+autoplot(rr, type = "prediction")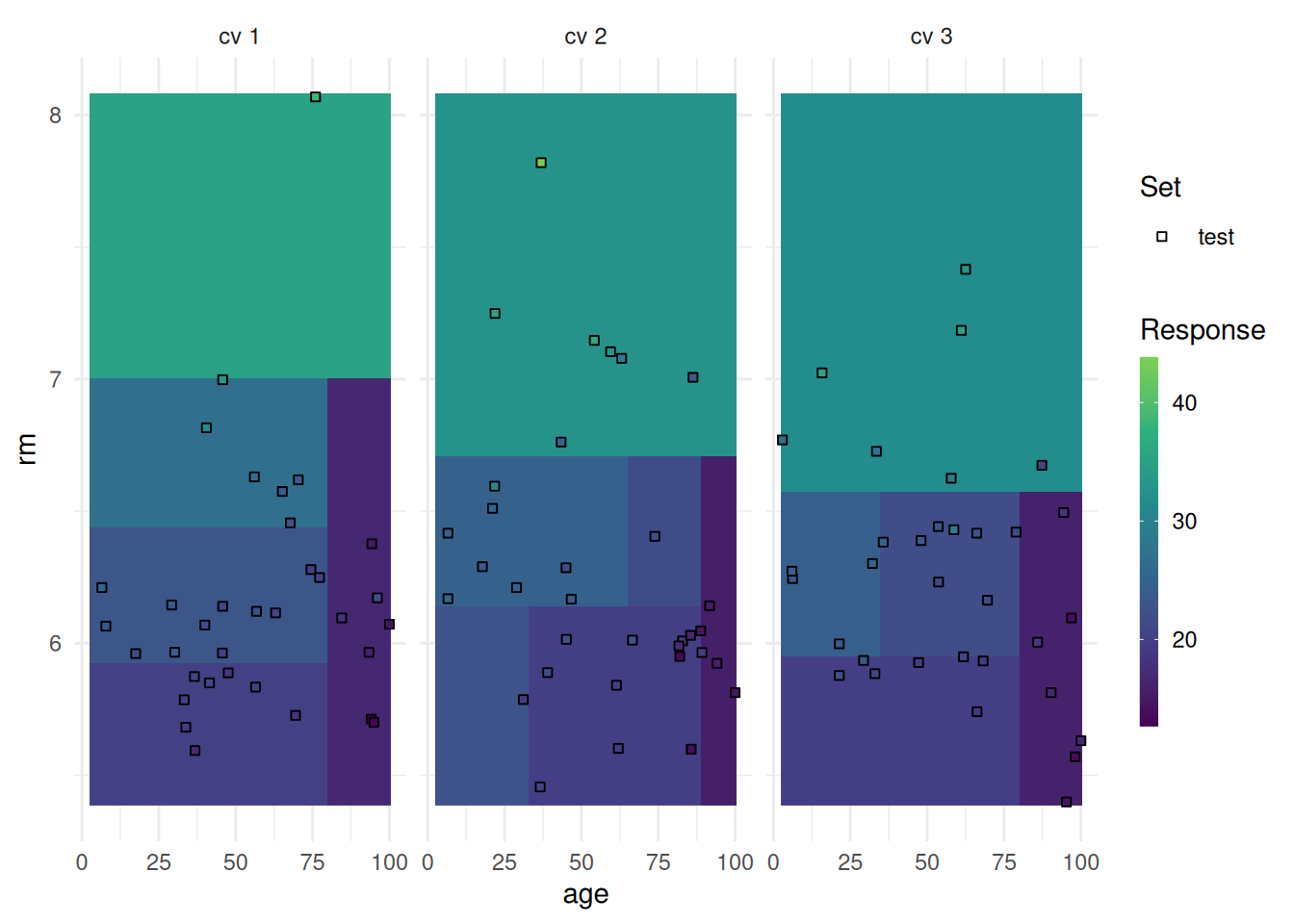
We show the performance distribution of a benchmark with multiple tasks.
+tasks = tsks(c("pima", "sonar"))
+learner = lrns(c("classif.featureless", "classif.rpart", "classif.xgboost"), predict_type = "prob")
+resampling = rsmps("cv")
+bmr = benchmark(benchmark_grid(tasks, learner, resampling))
+
+autoplot(bmr, type = "boxplot")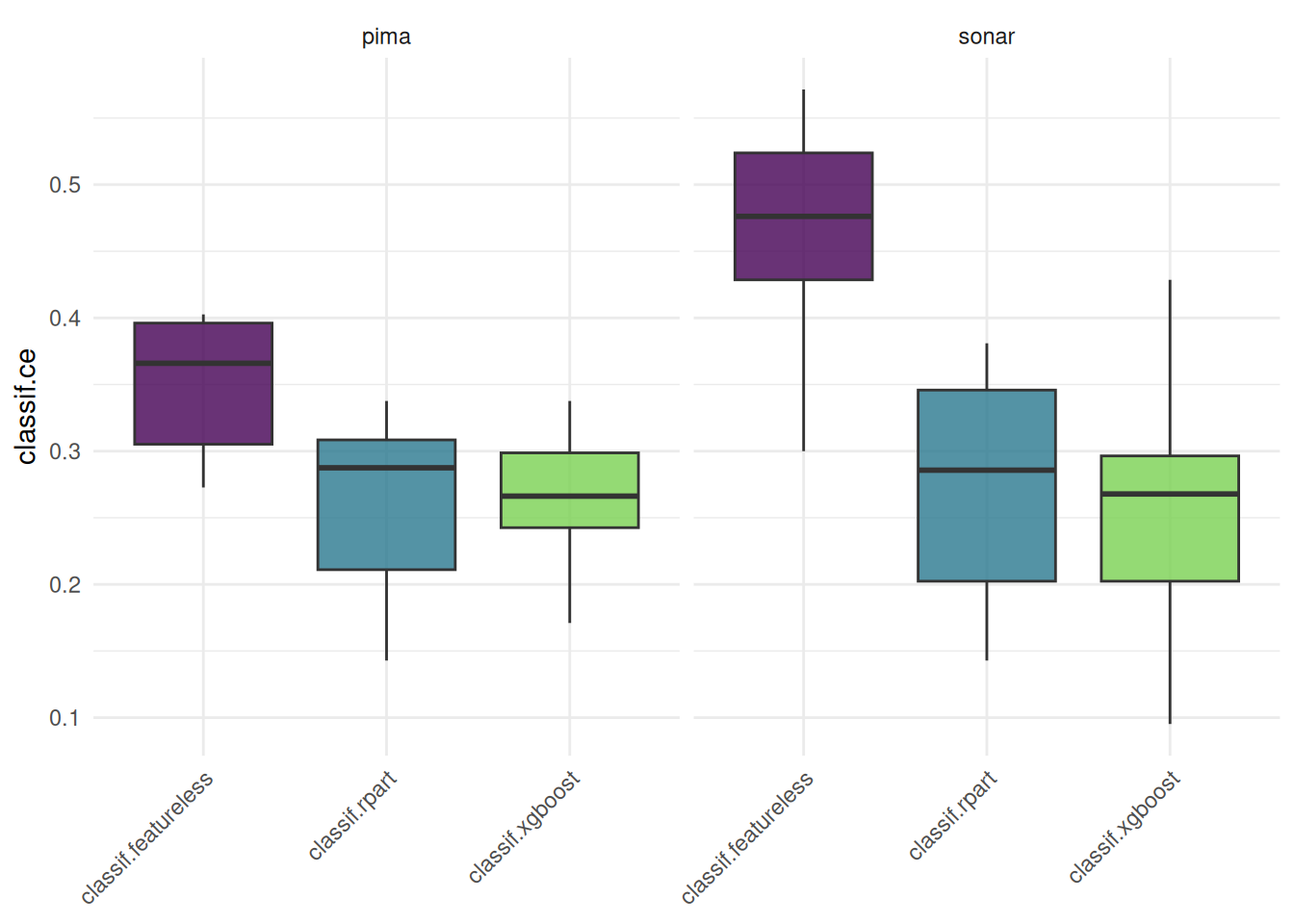
We plot a benchmark result with one task and multiple learners.
+tasks = tsk("pima")
+learner = lrns(c("classif.featureless", "classif.rpart", "classif.xgboost"), predict_type = "prob")
+resampling = rsmps("cv")
+bmr = benchmark(benchmark_grid(tasks, learner, resampling))We plot an roc curve for each learner.
+autoplot(bmr, type = "roc")
Alternatively, we can plot precision-recall curves.
+autoplot(bmr, type = "prc")
We tune the hyperparameters of a decision tree on the sonar task. The "performance" plot shows the performance over batches.
library(mlr3tuning)
+library(mlr3tuningspaces)
+library(mlr3learners)
+
+instance = tune(
+ tuner = tnr("gensa"),
+ task = tsk("sonar"),
+ learner = lts(lrn("classif.rpart")),
+ resampling = rsmp("holdout"),
+ measures = msr("classif.ce"),
+ term_evals = 100
+)
+
+autoplot(instance, type = "performance")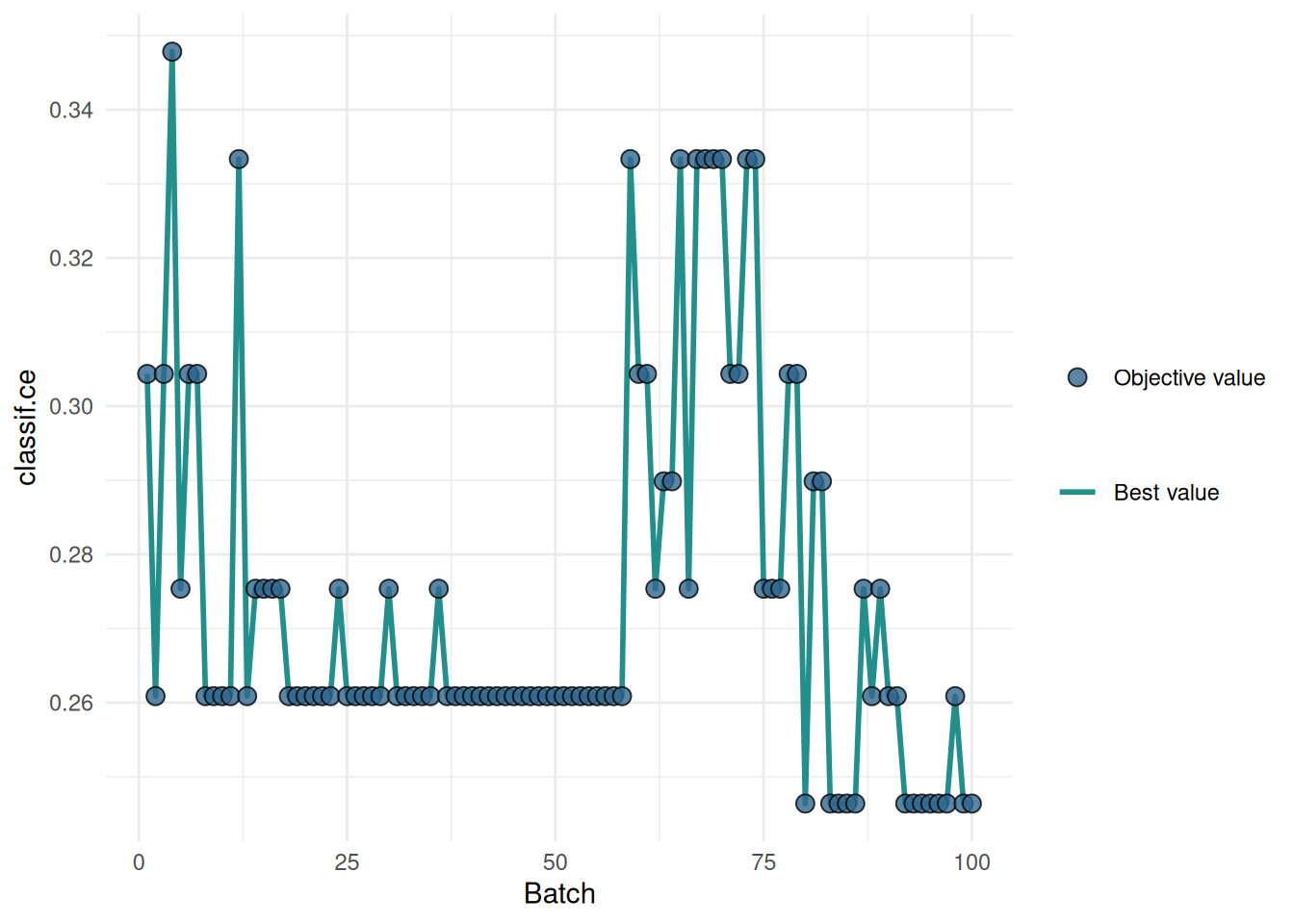
The "incumbent" plot shows the performance of the best hyperparameter setting over the number of evaluations.
autoplot(instance, type = "incumbent")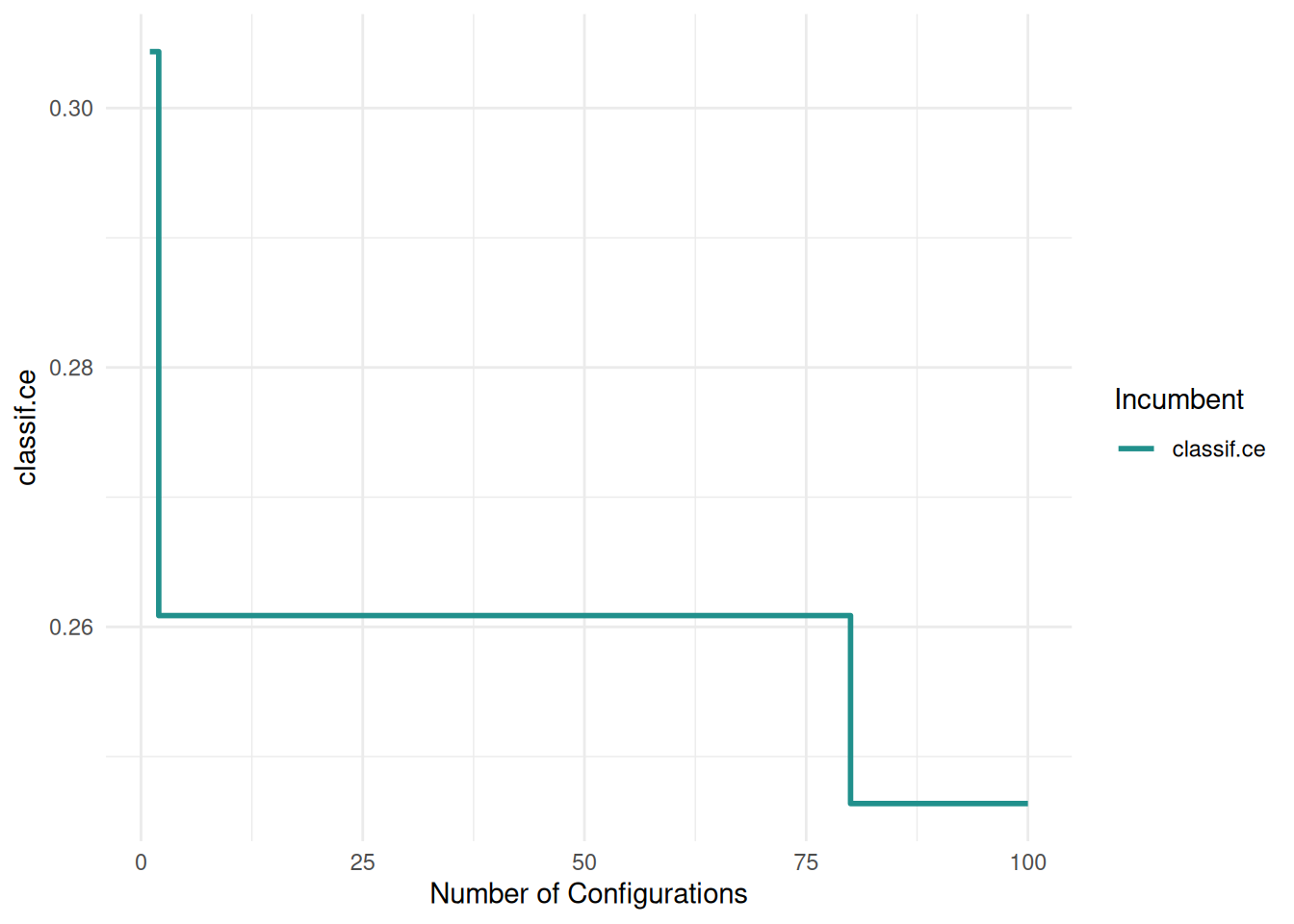
The "parameter" plot shows the performance for each hyperparameter setting.
autoplot(instance, type = "parameter", cols_x = c("cp", "minsplit"))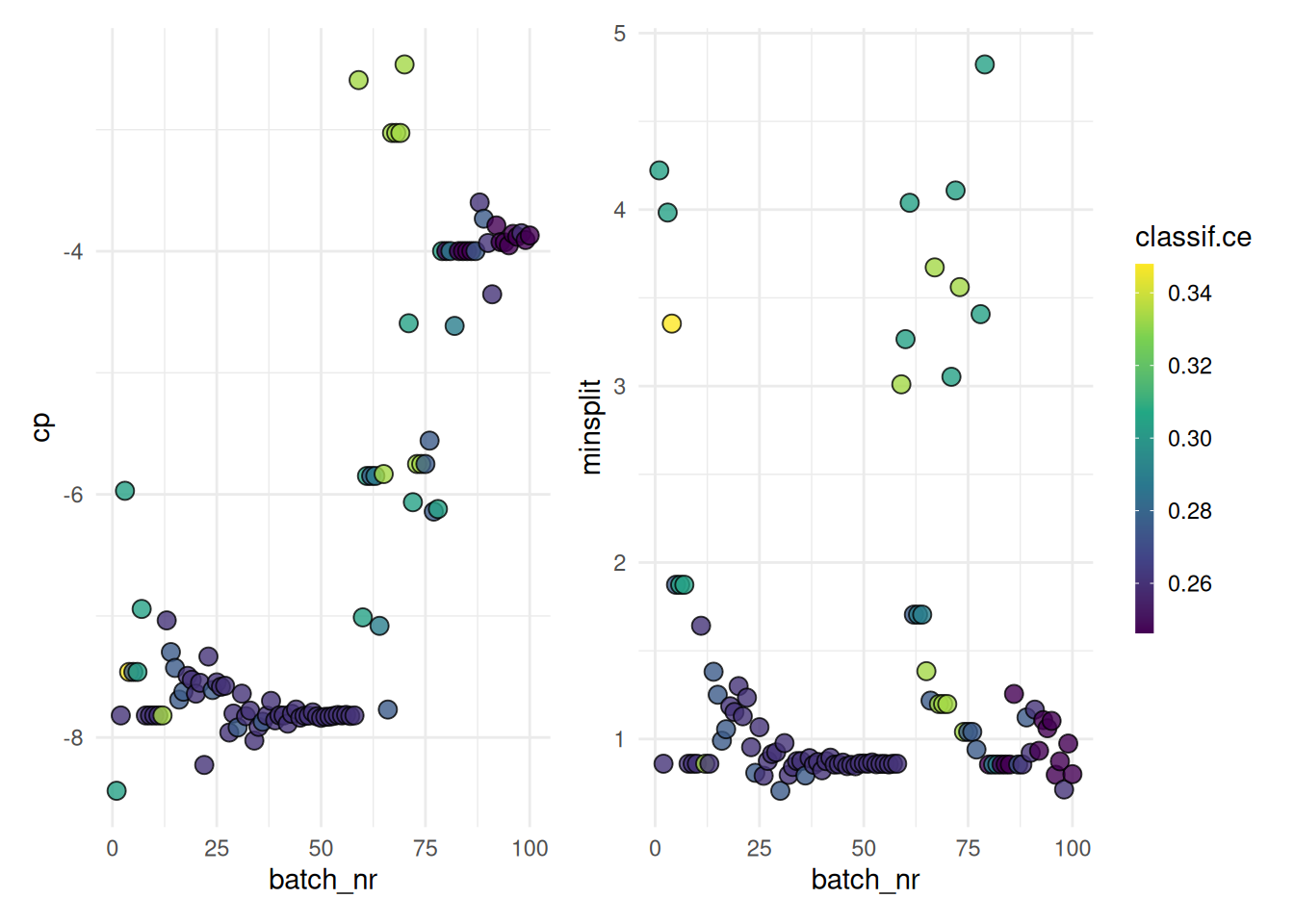
The "marginal" plot shows the performance of different hyperparameter values. The color indicates the batch.
autoplot(instance, type = "marginal", cols_x = "cp")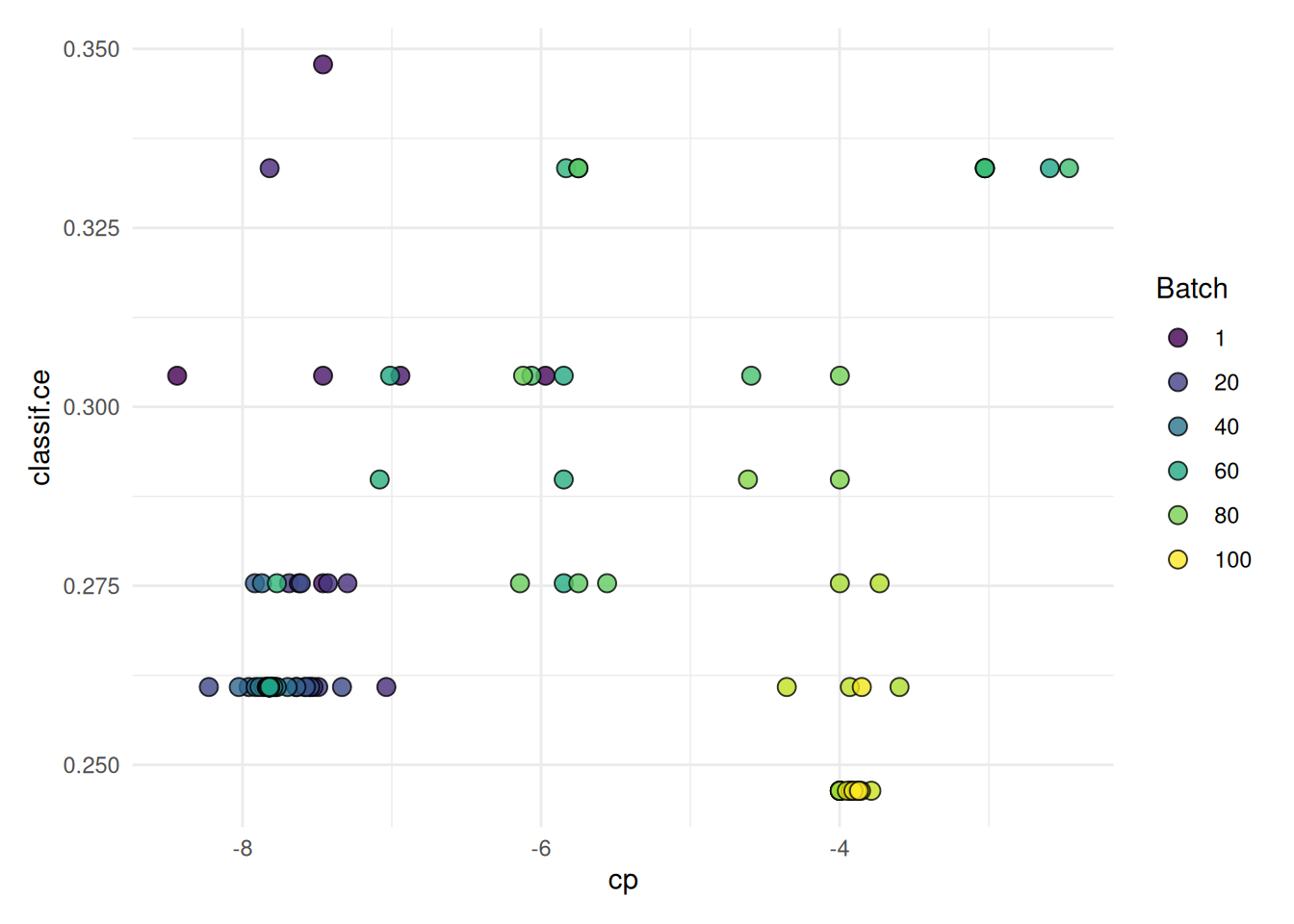
The "parallel" plot visualizes the relationship of hyperparameters.
autoplot(instance, type = "parallel")
We plot cp against minsplit and color the points by the performance.
autoplot(instance, type = "points", cols_x = c("cp", "minsplit"))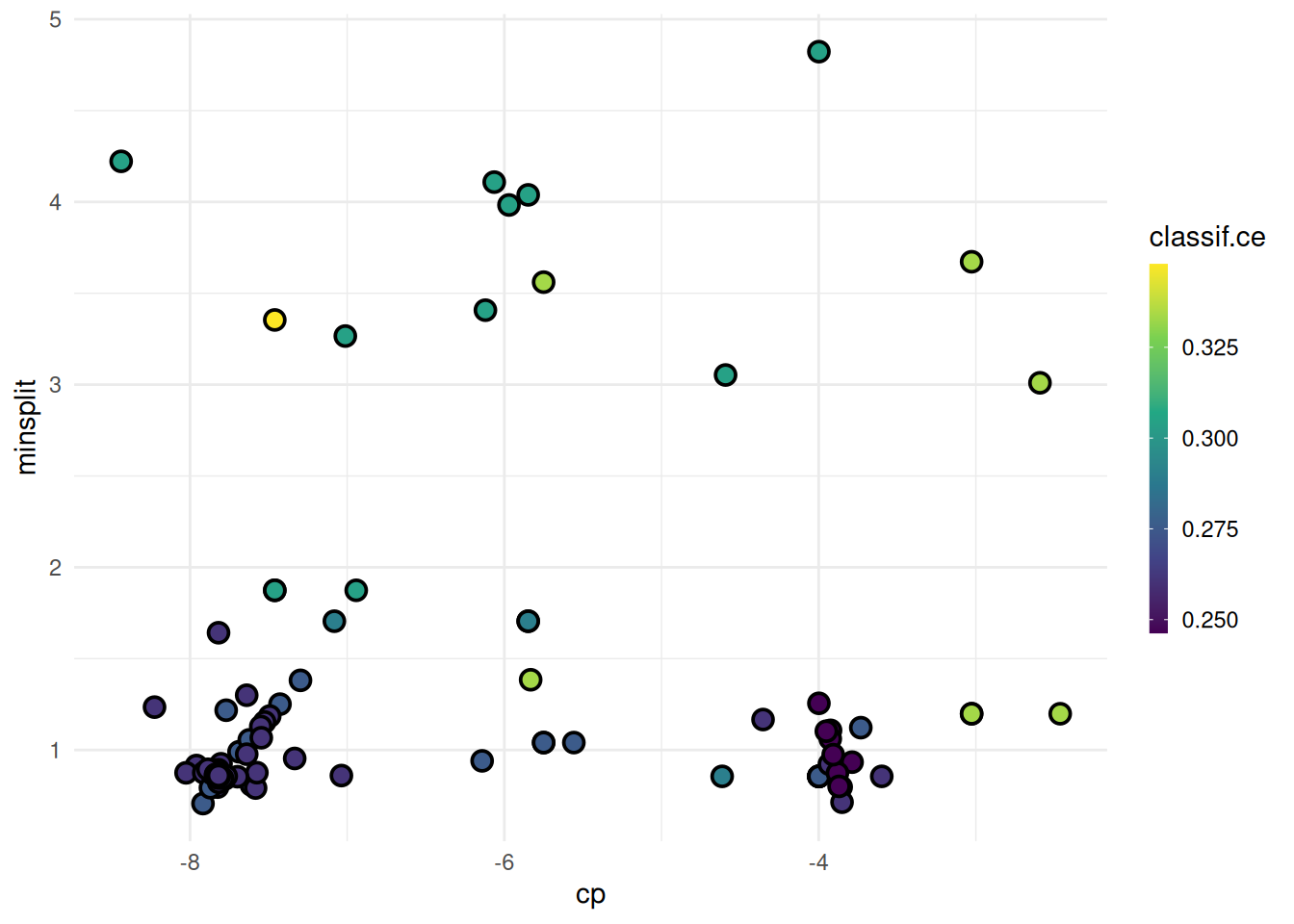
Next, we plot all hyperparameters against each other.
+autoplot(instance, type = "pairs")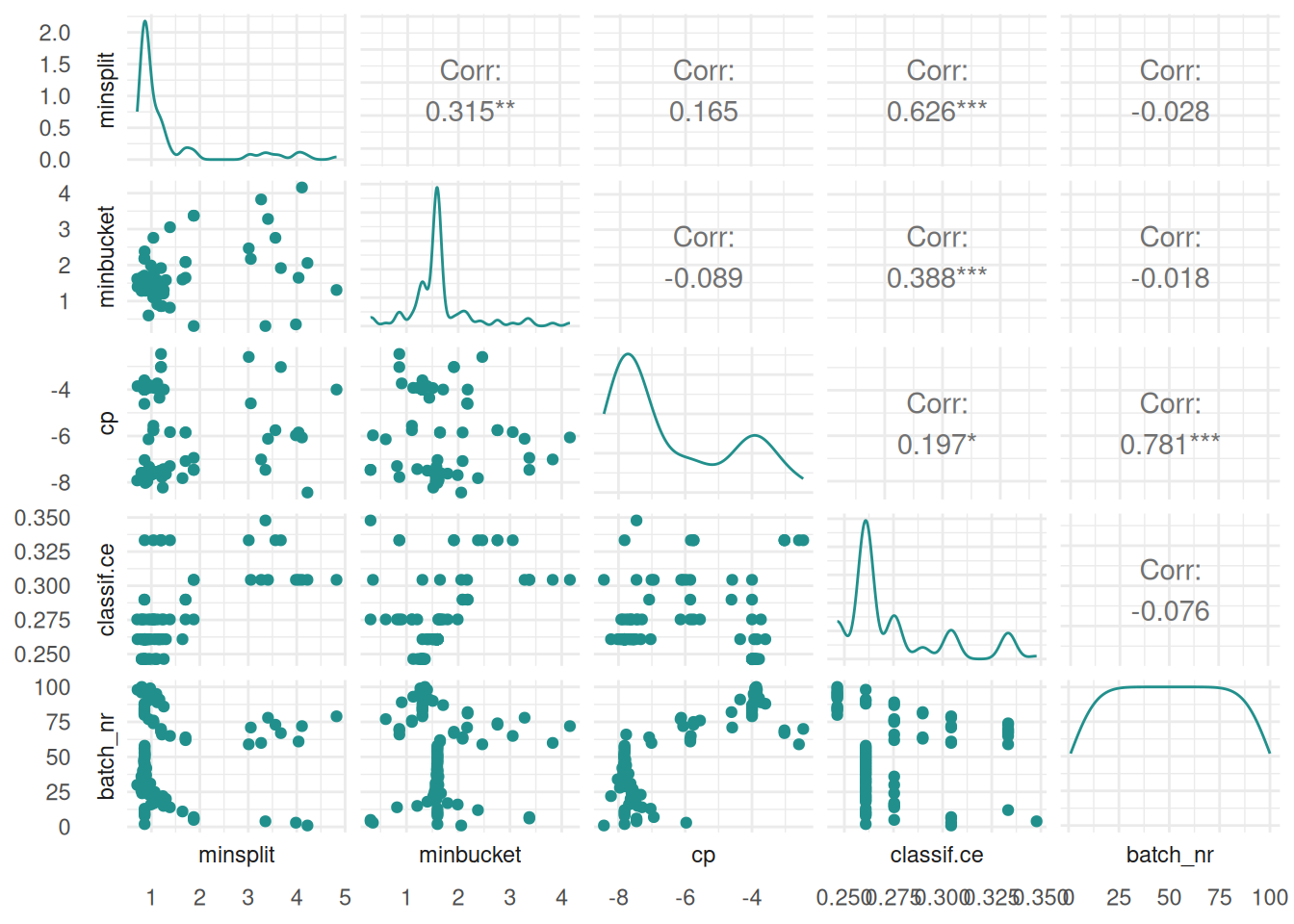
We plot the performance surface of two hyperparameters. The surface is interpolated with a learner.
+autoplot(instance, type = "surface", cols_x = c("cp", "minsplit"), learner = mlr3::lrn("regr.ranger"))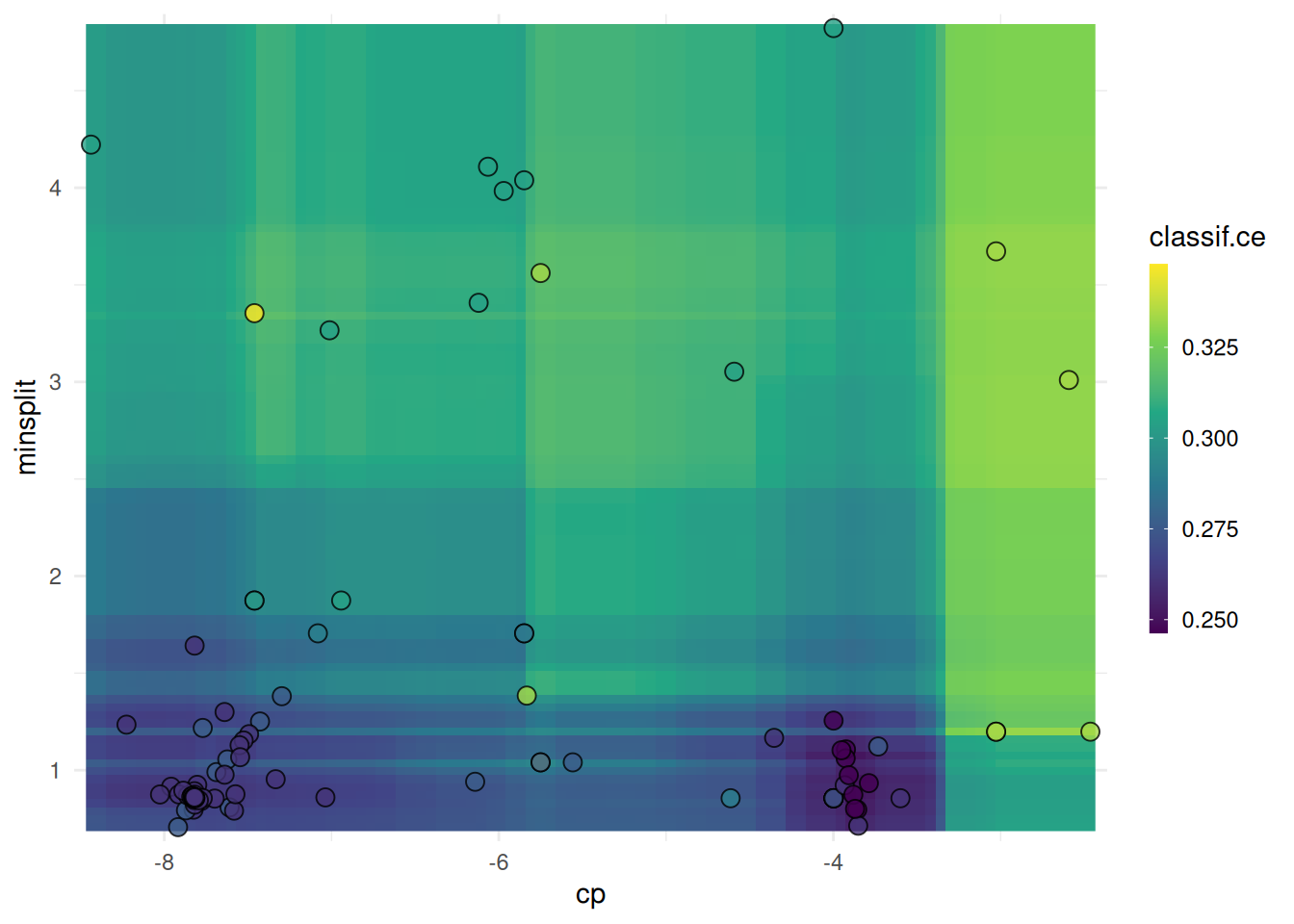
We plot filter scores for the mtcars task.
+library(mlr3filters)
+
+task = tsk("mtcars")
+f = flt("correlation")
+f$calculate(task)
+
+autoplot(f, n = 5)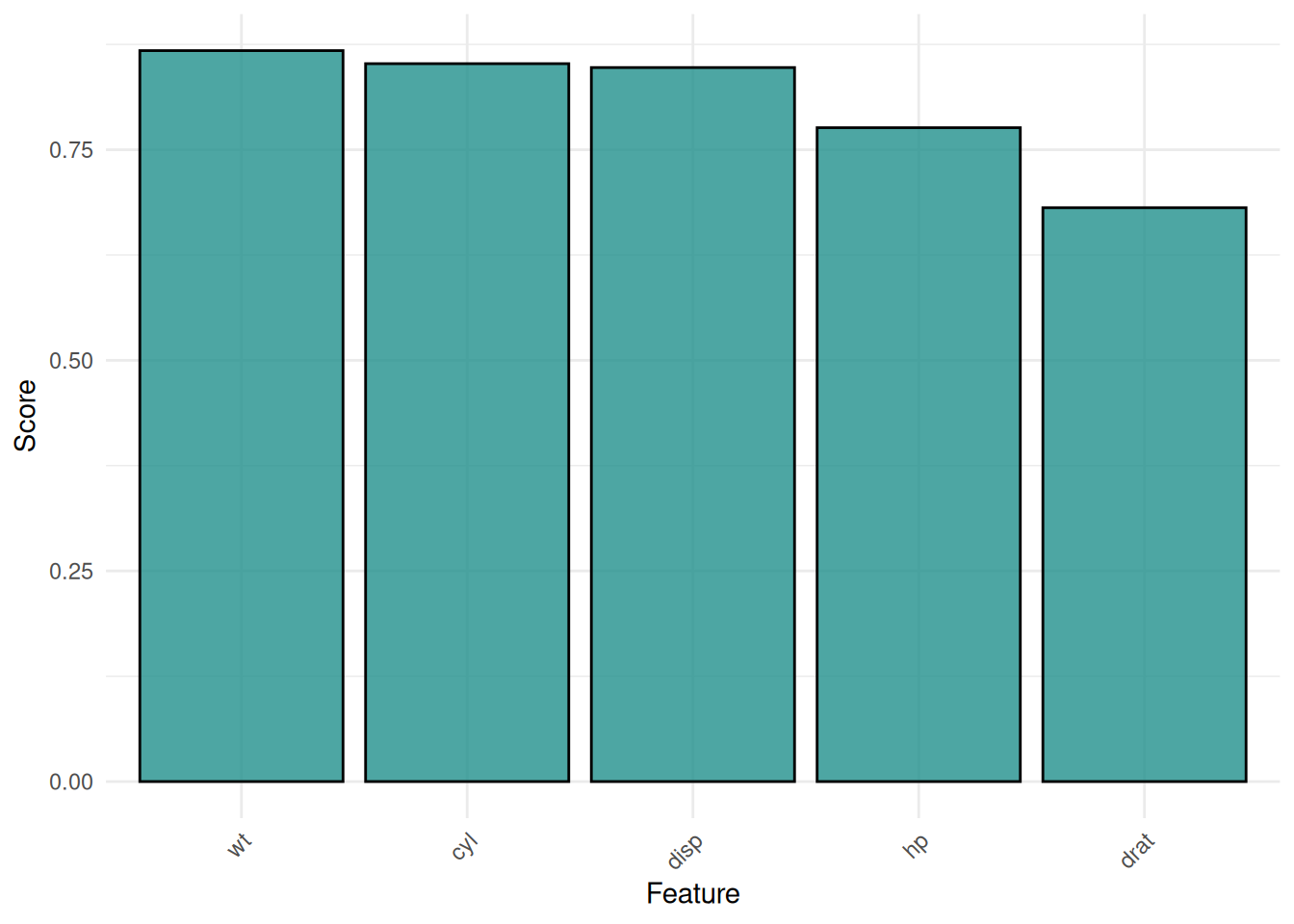
The mlr3viz package brings together the visualization functions of the mlr3 ecosystem. All plots are drawn with the autoplot() function and the appearance can be customized with the theme argument. If you need to highly customize a plot e.g. for a publication, we encourage you to check our code on GitHub. The code should be easily adaptable to your needs. We are also looking forward to new visualizations. You can suggest new plots in an issue on GitHub.
In this post, we optimize the hyperparameters of a simple classification tree on the Palmer Penguins data set with only a few lines of code.
First, we introduce tuning spaces and show the importance of transformation functions. Next, we execute the tuning and present the basic building blocks of tuning in mlr3. Finally, we fit a classification tree with optimized hyperparameters on the full data set.
+ +We load the mlr3verse package which pulls the most important packages for this example. Among other packages, it loads the hyperparameter optimization package of the mlr3 ecosystem mlr3tuning.
+library(mlr3verse)In this example, we use the Palmer Penguins data set which classifies 344 penguins in three species. The data set was collected from 3 islands in the Palmer Archipelago in Antarctica. It includes the name of the island, the size (flipper length, body mass, and bill dimension), and the sex of the penguin.
tsk("penguins")<TaskClassif:penguins> (344 x 8): Palmer Penguins
+* Target: species
+* Properties: multiclass
+* Features (7):
+ - int (3): body_mass, flipper_length, year
+ - dbl (2): bill_depth, bill_length
+ - fct (2): island, sexlibrary(palmerpenguins)
+library(ggplot2)
+ggplot(data = penguins, aes(x = flipper_length_mm, y = bill_length_mm)) +
+ geom_point(aes(color = species, shape = species), size = 3, alpha = 0.8) +
+ geom_smooth(method = "lm", se = FALSE, aes(color = species)) +
+ theme_minimal() +
+ scale_color_manual(values = c("darkorange","purple","cyan4")) +
+ labs(x = "Flipper length (mm)", y = "Bill length (mm)", color = "Penguin species", shape = "Penguin species") +
+ theme(
+ legend.position = c(0.85, 0.15),
+ legend.background = element_rect(fill = "white", color = NA),
+ text = element_text(size = 10))
We use the rpart classification tree. A learner stores all information about its hyperparameters in the slot $param_set. Not all parameters are tunable. We have to choose a subset of the hyperparameters we want to tune.
learner = lrn("classif.rpart")
+as.data.table(learner$param_set)[, list(id, class, lower, upper, nlevels)] id class lower upper nlevels
+ <char> <char> <num> <num> <num>
+ 1: cp ParamDbl 0 1 Inf
+ 2: keep_model ParamLgl NA NA 2
+ 3: maxcompete ParamInt 0 Inf Inf
+ 4: maxdepth ParamInt 1 30 30
+ 5: maxsurrogate ParamInt 0 Inf Inf
+ 6: minbucket ParamInt 1 Inf Inf
+ 7: minsplit ParamInt 1 Inf Inf
+ 8: surrogatestyle ParamInt 0 1 2
+ 9: usesurrogate ParamInt 0 2 3
+10: xval ParamInt 0 Inf InfThe package mlr3tuningspaces is a collection of search spaces for hyperparameter tuning from peer-reviewed articles. We use the search space from the Bischl et al. (2021) article.
+lts("classif.rpart.default")<TuningSpace:classif.rpart.default>: Classification Rpart with Default
+ id lower upper levels logscale
+ <char> <num> <num> <list> <lgcl>
+1: minsplit 2e+00 128.0 TRUE
+2: minbucket 1e+00 64.0 TRUE
+3: cp 1e-04 0.1 TRUEThe classification tree is mainly influenced by three hyperparameters:
+cp that controls when the learner considers introducing another branch.minsplit hyperparameter that controls how many observations must be present in a leaf for another split to be attempted.minbucket hyperparameter that the minimum number of observations in any terminal node.We argument the learner with the search space in one go.
+lts(lrn("classif.rpart"))<LearnerClassifRpart:classif.rpart>: Classification Tree
+* Model: -
+* Parameters: xval=0, minsplit=<RangeTuneToken>, minbucket=<RangeTuneToken>, cp=<RangeTuneToken>
+* Packages: mlr3, rpart
+* Predict Types: [response], prob
+* Feature Types: logical, integer, numeric, factor, ordered
+* Properties: importance, missings, multiclass, selected_features, twoclass, weightsThe column logscale indicates that the hyperparameters are tuned on the logarithmic scale. The tuning algorithm proposes hyperparameter values that are transformed with the exponential function before they are passed to the learner. For example, the cp parameter is bounded between 0 and 1. The tuning algorithm searches between log(1e-04) and log(1e-01) but the learner gets the transformed values between 1e-04 and 1e-01. Using the log transformation emphasizes smaller cp values but also creates large values.
lts("classif.rpart.default")<TuningSpace:classif.rpart.default>: Classification Rpart with Default
+ id lower upper levels logscale
+ <char> <num> <num> <list> <lgcl>
+1: minsplit 2e+00 128.0 TRUE
+2: minbucket 1e+00 64.0 TRUE
+3: cp 1e-04 0.1 TRUEThe tune() function controls and executes the tuning. The method sets the optimization algorithm. The mlr3 ecosystem offers various optimization algorithms e.g. Random Search, GenSA, and Hyperband. In this example, we will use a simple grid search with a grid resolution of 5. Our three-dimensional grid consists of hyperparameter configurations. The
resampling strategy and performance measure specify how the performance of a model is evaluated. We choose a 3-fold cross-validation and use the classification error.
instance = tune(
+ tuner = tnr("grid_search", resolution = 5),
+ task = tsk("penguins"),
+ learner = lts(lrn("classif.rpart")),
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce")
+)The tune() function returns a tuning instance that includes an archive with all evaluated hyperparameter configurations.
as.data.table(instance$archive)[, list(minsplit, minbucket, cp, classif.ce, resample_result)] minsplit minbucket cp classif.ce resample_result
+ <num> <num> <num> <num> <list>
+ 1: 2.7764798 2.087194 -2.302585 0.06107297 <ResampleResult[21]>
+ 2: 1.7348135 4.174387 -2.302585 0.18001526 <ResampleResult[21]>
+ 3: 0.6931472 1.043597 -9.210340 0.02906178 <ResampleResult[21]>
+ 4: 0.6931472 3.130790 -9.210340 0.06107297 <ResampleResult[21]>
+ 5: 2.7764798 3.130790 -4.029524 0.06107297 <ResampleResult[21]>
+ ---
+121: 3.8181461 0.000000 -5.756463 0.04065599 <ResampleResult[21]>
+122: 0.6931472 0.000000 -7.483402 0.02616323 <ResampleResult[21]>
+123: 0.6931472 3.130790 -2.302585 0.06107297 <ResampleResult[21]>
+124: 1.7348135 2.087194 -4.029524 0.04937707 <ResampleResult[21]>
+125: 3.8181461 0.000000 -2.302585 0.06107297 <ResampleResult[21]>The best configuration and the corresponding measured performance can be retrieved from the tuning instance.
+instance$result minsplit minbucket cp learner_param_vals x_domain classif.ce
+ <num> <num> <num> <list> <list> <num>
+1: 0.6931472 0 -9.21034 <list[4]> <list[3]> 0.02616323The $result_learner_param_vals field contains the best hyperparameter setting on the learner scale.
instance$result_learner_param_vals$xval
+[1] 0
+
+$minsplit
+[1] 2
+
+$minbucket
+[1] 1
+
+$cp
+[1] 1e-04The learner we use to make predictions on new data is called the final model. The final model is trained on the full data set. We add the optimized hyperparameters to the learner and train the learner on the full dataset.
+learner = lrn("classif.rpart")
+learner$param_set$values = instance$result_learner_param_vals
+learner$train(tsk("penguins"))The trained model can now be used to predict new, external data.
+ + + +In this post, we use early stopping to reduce overfitting when training an XGBoost model. We start with a short recap on early stopping and overfitting. After that, we use the early stopping mechanism of XGBoost and train a model on the Spam Classification data set. Finally we show how to simultaneously tune hyperparameters and use early stopping. The reader should be familiar with tuning in the mlr3 ecosystem.
Early stopping is a technique used to reduce overfitting when fitting a model in an iterative process. Overfitting occurs when a model fits increasingly to the training data but the performance on unseen data decreases. This means the model’s training error decreases, while its test performance deteriorates. When using early stopping, the performance is monitored on a test set, and the training stops when performance decreases in a specific number of iterations.
+We initialize the random number generator with a fixed seed for reproducibility. The mlr3verse package provides all functions required for this example.
+set.seed(7832)
+
+library(mlr3verse)When training an XGBoost model, we can use early stopping to find the optimal number of boosting rounds. The partition() function splits the observations of the task into two disjoint sets. We use 80% of observations to train the model and the remaining 20% as the test set to monitor the performance.
task = tsk("spam")
+split = partition(task, ratio = 0.8)
+task$set_row_roles(split$test, "test")The early_stopping_set parameter controls which set is used to monitor the performance. Additionally, we need to define the range in which the performance must increase with early_stopping_rounds and the maximum number of boosting rounds with nrounds. In this example, the training is stopped when the classification error is not decreasing for 100 rounds or 1000 rounds are reached.
learner = lrn("classif.xgboost",
+ nrounds = 1000,
+ early_stopping_rounds = 100,
+ early_stopping_set = "test",
+ eval_metric = "error"
+)We train the learner with early stopping.
+learner$train(task)The $evaluation_log of the model stores the performance scores on the training and test set. Figure 1 shows that the classification error on the training set decreases, whereas the error on the test set increases after 20 rounds.
library(ggplot2)
+library(data.table)
+
+data = melt(
+ learner$model$evaluation_log,
+ id.vars = "iter",
+ variable.name = "set",
+ value.name = "error"
+)
+
+ggplot(data, aes(x = iter, y = error, group = set)) +
+ geom_line(aes(color = set)) +
+ geom_vline(aes(xintercept = learner$model$best_iteration), color = "grey") +
+ scale_colour_manual(values=c("#f8766d", "#00b0f6"), labels = c("Train", "Test")) +
+ labs(x = "Rounds", y = "Classification Error", color = "Set") +
+ theme_minimal()
The slot $best_iteration contains the optimal number of boosting rounds.
learner$model$best_iteration[1] 20Note that, learner$predict() will use the model from the last iteration, not the best one. See the next section on how to fit a model with the optimal number of boosting rounds and hyperparameter configuration.
In this section, we want to tune the hyperparameters of an XGBoost model and find the optimal number of boosting rounds in one go. For this, we need the early stopping callback which handles early stopping during the tuning process. The performance of a hyperparameter configuration is evaluated with a resampling strategy while tuning e.g. 3-fold cross-validation. In each resampling iteration, a new XGBoost model is trained and early stopping is used to find the optimal number of boosting rounds. This results in three different optimal numbers of boosting rounds for one hyperparameter configuration when applying 3-fold cross-validation. The callback picks the maximum of the three values and writes it to the archive. It uses the maximum value because the final model is fitted on the complete data set. Now let’s start with a practical example.
First, we load the XGBoost learner and set the early stopping parameters.
+learner = lrn("classif.xgboost",
+ nrounds = 1000,
+ early_stopping_rounds = 100,
+ early_stopping_set = "test"
+)Next, we load a predefined tuning space from the mlr3tuningspaces package. The tuning space includes the most commonly tuned parameters of XGBoost.
+tuning_space = lts("classif.xgboost.default")
+as.data.table(tuning_space) id lower upper logscale
+1: eta 1e-04 1 TRUE
+2: nrounds 1e+00 5000 FALSE
+3: max_depth 1e+00 20 FALSE
+4: colsample_bytree 1e-01 1 FALSE
+5: colsample_bylevel 1e-01 1 FALSE
+6: lambda 1e-03 1000 TRUE
+7: alpha 1e-03 1000 TRUE
+8: subsample 1e-01 1 FALSEWe argument the learner with the tuning space.
+learner = lts(learner)The default tuning space contains the nrounds hyperparameter. We have to overwrite it with an upper bound for early stopping.
learner$param_set$set_values(nrounds = 1000)We run a small batch of random hyperparameter configurations.
+instance = tune(
+ tuner = tnr("random_search", batch_size = 2),
+ task = task,
+ learner = learner,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ term_evals = 4,
+ callbacks = clbk("mlr3tuning.early_stopping")
+)We can see that the optimal number of boosting rounds (max_nrounds) strongly depends on the other hyperparameters.
as.data.table(instance$archive)[, list(batch_nr, max_nrounds, eta, max_depth, colsample_bytree, colsample_bylevel, lambda, alpha, subsample)] batch_nr max_nrounds eta max_depth colsample_bytree colsample_bylevel lambda alpha subsample
+1: 1 1000 -4.129996 13 0.5466492 0.3051989 -0.9448420 2.535477 0.5454539
+2: 1 998 -3.338899 8 0.6193478 0.7392354 -2.2338126 1.793921 0.2294161
+3: 2 1000 -6.059897 5 0.6118892 0.5445475 -6.5698270 5.414224 0.9635730
+4: 2 1000 -4.528129 1 0.1094186 0.2526143 -0.7535105 -3.041829 0.5934376In the best hyperparameter configuration, the value of nrounds is replaced by max_nrounds and early stopping is deactivated.
instance$result_learner_param_vals$nrounds
+[1] 998
+
+$nthread
+[1] 1
+
+$verbose
+[1] 0
+
+$early_stopping_set
+[1] "none"
+
+$eta
+[1] 0.03547598
+
+$max_depth
+[1] 8
+
+$colsample_bytree
+[1] 0.6193478
+
+$colsample_bylevel
+[1] 0.7392354
+
+$lambda
+[1] 0.1071192
+
+$alpha
+[1] 6.012984
+
+$subsample
+[1] 0.2294161Finally, fit the final model on the complete data set.
+learner = lrn("classif.xgboost")
+learner$param_set$values = instance$result_learner_param_vals
+learner$train(task)The trained model can now be used to make predictions on new data.
+We can also use the AutoTuner to get a tuned XGBoost model. Note that, early stopping is deactivated when the final model is fitted.
The package mlr3tuningspaces offers a selection of published search spaces for many popular machine learning algorithms. In this post, we show how to tune a mlr3 learners with these search spaces.
The packages mlr3verse and mlr3tuningspaces are required for this demonstration:
+library(mlr3verse)
+library(mlr3tuningspaces)We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented.
+set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")In the example, we use the pima indian diabetes data set which is used to predict whether or not a patient has diabetes. The patients are characterized by 8 numeric features, some of them have missing values.
# retrieve the task from mlr3
+task = tsk("pima")
+
+# generate a quick textual overview using the skimr package
+skimr::skim(task$data())| Name | +task$data() | +
| Number of rows | +768 | +
| Number of columns | +9 | +
| Key | +NULL | +
| _______________________ | ++ |
| Column type frequency: | ++ |
| factor | +1 | +
| numeric | +8 | +
| ________________________ | ++ |
| Group variables | +None | +
Variable type: factor
+| skim_variable | +n_missing | +complete_rate | +ordered | +n_unique | +top_counts | +
|---|---|---|---|---|---|
| diabetes | +0 | +1 | +FALSE | +2 | +neg: 500, pos: 268 | +
Variable type: numeric
+| skim_variable | +n_missing | +complete_rate | +mean | +sd | +p0 | +p25 | +p50 | +p75 | +p100 | +hist | +
|---|---|---|---|---|---|---|---|---|---|---|
| age | +0 | +1.00 | +33.24 | +11.76 | +21.00 | +24.00 | +29.00 | +41.00 | +81.00 | +▇▃▁▁▁ | +
| glucose | +5 | +0.99 | +121.69 | +30.54 | +44.00 | +99.00 | +117.00 | +141.00 | +199.00 | +▁▇▇▃▂ | +
| insulin | +374 | +0.51 | +155.55 | +118.78 | +14.00 | +76.25 | +125.00 | +190.00 | +846.00 | +▇▂▁▁▁ | +
| mass | +11 | +0.99 | +32.46 | +6.92 | +18.20 | +27.50 | +32.30 | +36.60 | +67.10 | +▅▇▃▁▁ | +
| pedigree | +0 | +1.00 | +0.47 | +0.33 | +0.08 | +0.24 | +0.37 | +0.63 | +2.42 | +▇▃▁▁▁ | +
| pregnant | +0 | +1.00 | +3.85 | +3.37 | +0.00 | +1.00 | +3.00 | +6.00 | +17.00 | +▇▃▂▁▁ | +
| pressure | +35 | +0.95 | +72.41 | +12.38 | +24.00 | +64.00 | +72.00 | +80.00 | +122.00 | +▁▃▇▂▁ | +
| triceps | +227 | +0.70 | +29.15 | +10.48 | +7.00 | +22.00 | +29.00 | +36.00 | +99.00 | +▆▇▁▁▁ | +
For tuning, it is important to create a search space that defines the type and range of the hyperparameters. A learner stores all information about its hyperparameters in the slot $param_set. Usually, we have to chose a subset of hyperparameters we want to tune.
lrn("classif.rpart")$param_set<ParamSet>
+ id class lower upper nlevels default value
+ 1: cp ParamDbl 0 1 Inf 0.01
+ 2: keep_model ParamLgl NA NA 2 FALSE
+ 3: maxcompete ParamInt 0 Inf Inf 4
+ 4: maxdepth ParamInt 1 30 30 30
+ 5: maxsurrogate ParamInt 0 Inf Inf 5
+ 6: minbucket ParamInt 1 Inf Inf <NoDefault[3]>
+ 7: minsplit ParamInt 1 Inf Inf 20
+ 8: surrogatestyle ParamInt 0 1 2 0
+ 9: usesurrogate ParamInt 0 2 3 2
+10: xval ParamInt 0 Inf Inf 10 0At the heart of mlr3tuningspaces is the R6 class TuningSpace. It stores a list of TuneToken, helper functions and additional meta information. The list of TuneToken can be directly applied to the $values slot of a learner’s ParamSet. The search spaces are stored in the mlr_tuning_spaces dictionary.
as.data.table(mlr_tuning_spaces) key label learner n_values
+ 1: classif.glmnet.default Classification GLM with Default classif.glmnet 2
+ 2: classif.glmnet.rbv1 Classification GLM with RandomBot classif.glmnet 2
+ 3: classif.glmnet.rbv2 Classification GLM with RandomBot classif.glmnet 2
+ 4: classif.kknn.default Classification KKNN with Default classif.kknn 3
+ 5: classif.kknn.rbv1 Classification KKNN with RandomBot classif.kknn 1
+ 6: classif.kknn.rbv2 Classification KKNN with RandomBot classif.kknn 1
+ 7: classif.ranger.default Classification Ranger with Default classif.ranger 4
+ 8: classif.ranger.rbv1 Classification Ranger with RandomBot classif.ranger 6
+ 9: classif.ranger.rbv2 Classification Ranger with RandomBot classif.ranger 8
+10: classif.rpart.default Classification Rpart with Default classif.rpart 3
+11: classif.rpart.rbv1 Classification Rpart with RandomBot classif.rpart 4
+12: classif.rpart.rbv2 Classification Rpart with RandomBot classif.rpart 4
+13: classif.svm.default Classification SVM with Default classif.svm 4
+14: classif.svm.rbv1 Classification SVM with RandomBot classif.svm 4
+15: classif.svm.rbv2 Classification SVM with RandomBot classif.svm 5
+16: classif.xgboost.default Classification XGBoost with Default classif.xgboost 8
+17: classif.xgboost.rbv1 Classification XGBoost with RandomBot classif.xgboost 10
+18: classif.xgboost.rbv2 Classification XGBoost with RandomBot classif.xgboost 13
+19: regr.glmnet.default Regression GLM with Default regr.glmnet 2
+20: regr.glmnet.rbv1 Regression GLM with RandomBot regr.glmnet 2
+21: regr.glmnet.rbv2 Regression GLM with RandomBot regr.glmnet 2
+22: regr.kknn.default Regression KKNN with Default regr.kknn 3
+23: regr.kknn.rbv1 Regression KKNN with RandomBot regr.kknn 1
+24: regr.kknn.rbv2 Regression KKNN with RandomBot regr.kknn 1
+25: regr.ranger.default Regression Ranger with Default regr.ranger 4
+26: regr.ranger.rbv1 Regression Ranger with RandomBot regr.ranger 6
+27: regr.ranger.rbv2 Regression Ranger with RandomBot regr.ranger 7
+28: regr.rpart.default Regression Rpart with Default regr.rpart 3
+29: regr.rpart.rbv1 Regression Rpart with RandomBot regr.rpart 4
+30: regr.rpart.rbv2 Regression Rpart with RandomBot regr.rpart 4
+31: regr.svm.default Regression SVM with Default regr.svm 4
+32: regr.svm.rbv1 Regression SVM with RandomBot regr.svm 4
+33: regr.svm.rbv2 Regression SVM with RandomBot regr.svm 5
+34: regr.xgboost.default Regression XGBoost with Default regr.xgboost 8
+35: regr.xgboost.rbv1 Regression XGBoost with RandomBot regr.xgboost 10
+36: regr.xgboost.rbv2 Regression XGBoost with RandomBot regr.xgboost 13
+ key label learner n_valuesWe can use the sugar function lts() to retrieve a TuningSpace.
tuning_space_rpart = lts("classif.rpart.default")
+tuning_space_rpart<TuningSpace:classif.rpart.default>: Classification Rpart with Default
+ id lower upper levels logscale
+1: minsplit 2e+00 128.0 TRUE
+2: minbucket 1e+00 64.0 TRUE
+3: cp 1e-04 0.1 TRUEThe $values slot contains the list of of TuneToken.
tuning_space_rpart$values$minsplit
+Tuning over:
+range [2, 128] (log scale)
+
+
+$minbucket
+Tuning over:
+range [1, 64] (log scale)
+
+
+$cp
+Tuning over:
+range [1e-04, 0.1] (log scale)We apply the search space and tune the learner.
learner = lrn("classif.rpart")
+
+learner$param_set$values = tuning_space_rpart$values
+
+instance = tune(
+ tuner = tnr("random_search"),
+ task = tsk("pima"),
+ learner = learner,
+ resampling = rsmp ("holdout"),
+ measure = msr("classif.ce"),
+ term_evals = 10)
+
+instance$result minsplit minbucket cp learner_param_vals x_domain classif.ce
+1: 3.40059 1.963618 -4.114895 <list[3]> <list[3]> 0.2539062We can also get the learner with search space already applied from the TuningSpace.
learner = tuning_space_rpart$get_learner()
+print(learner$param_set)<ParamSet>
+ id class lower upper nlevels default value
+ 1: cp ParamDbl 0 1 Inf 0.01 <RangeTuneToken[2]>
+ 2: keep_model ParamLgl NA NA 2 FALSE
+ 3: maxcompete ParamInt 0 Inf Inf 4
+ 4: maxdepth ParamInt 1 30 30 30
+ 5: maxsurrogate ParamInt 0 Inf Inf 5
+ 6: minbucket ParamInt 1 Inf Inf <NoDefault[3]> <RangeTuneToken[2]>
+ 7: minsplit ParamInt 1 Inf Inf 20 <RangeTuneToken[2]>
+ 8: surrogatestyle ParamInt 0 1 2 0
+ 9: usesurrogate ParamInt 0 2 3 2
+10: xval ParamInt 0 Inf Inf 10 0This method also allows to set constant parameters.
+learner = tuning_space_rpart$get_learner(maxdepth = 15)
+print(learner$param_set)<ParamSet>
+ id class lower upper nlevels default value
+ 1: cp ParamDbl 0 1 Inf 0.01 <RangeTuneToken[2]>
+ 2: keep_model ParamLgl NA NA 2 FALSE
+ 3: maxcompete ParamInt 0 Inf Inf 4
+ 4: maxdepth ParamInt 1 30 30 30 15
+ 5: maxsurrogate ParamInt 0 Inf Inf 5
+ 6: minbucket ParamInt 1 Inf Inf <NoDefault[3]> <RangeTuneToken[2]>
+ 7: minsplit ParamInt 1 Inf Inf 20 <RangeTuneToken[2]>
+ 8: surrogatestyle ParamInt 0 1 2 0
+ 9: usesurrogate ParamInt 0 2 3 2
+10: xval ParamInt 0 Inf Inf 10 0The lts() function sets the default search space directly to a learner.
learner = lts(lrn("classif.rpart", maxdepth = 15))
+print(learner$param_set)<ParamSet>
+ id class lower upper nlevels default value
+ 1: cp ParamDbl 0 1 Inf 0.01 <RangeTuneToken[2]>
+ 2: keep_model ParamLgl NA NA 2 FALSE
+ 3: maxcompete ParamInt 0 Inf Inf 4
+ 4: maxdepth ParamInt 1 30 30 30 15
+ 5: maxsurrogate ParamInt 0 Inf Inf 5
+ 6: minbucket ParamInt 1 Inf Inf <NoDefault[3]> <RangeTuneToken[2]>
+ 7: minsplit ParamInt 1 Inf Inf 20 <RangeTuneToken[2]>
+ 8: surrogatestyle ParamInt 0 1 2 0
+ 9: usesurrogate ParamInt 0 2 3 2
+10: xval ParamInt 0 Inf Inf 10 0This is the fourth part of the practical tuning series. The other parts can be found here:
+In this post, we teach how to run various jobs in mlr3 in parallel. The goal is to map computational jobs (e.g. evaluation of one configuration) to a pool of workers (usually physical CPU cores, sometimes remote computational nodes) to reduce the run time needed for tuning.
+ +We load the mlr3verse package which pulls in the most important packages for this example. Additionally, make sure you have installed the packages future and future.apply.
+library(mlr3verse)We decrease the verbosity of the logger to keep the output clearly represented. The lgr package is used for logging in all mlr3 packages. The mlr3 logger prints the logging messages from the base package, whereas the bbotk logger is responsible for logging messages from the optimization packages (e.g. mlr3tuning ).
set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")The workers are specified by the parallel backend which orchestrates starting up, shutting down, and communication with the workers. On a single machine, multisession and multicore are common backends. The multisession backend spawns new background R processes. It is available on all platforms.
future::plan("multisession")The multicore backend uses forked R processes which allows the workers to access R objects in a shared memory. This reduces the overhead since R objects are only copied in memory if they are modified. Unfortunately, forking processes is not supported on Windows and when running R from within RStudio.
future::plan("multicore")Both backends support the workers argument that specifies the number of used cores.
Use this code if your code should run with the multicore backend when possible.
if (future::supportsMulticore()) {
+ future::plan(future::multicore)
+} else {
+ future::plan(future::multisession)
+}The resample() and benchmark() functions in mlr3 can be executed in parallel. The parallelization is triggered by simply declaring a plan via future::plan().
future::plan("multisession")
+
+task = tsk("pima")
+learner = lrn("classif.rpart") # classification tree
+resampling = rsmp("cv", folds = 3)
+
+resample(task, learner, resampling)<ResampleResult> with 3 resampling iterations
+ task_id learner_id resampling_id iteration warnings errors
+ pima classif.rpart cv 1 0 0
+ pima classif.rpart cv 2 0 0
+ pima classif.rpart cv 3 0 0The 3-fold cross-validation gives us 3 jobs since each resampling iteration is executed in parallel.
+The benchmark() function accepts a design of experiments as input where each experiment is defined as a combination of a task, a learner, and a resampling strategy. For each experiment, resampling is performed. The nested loop over experiments and resampling iterations is flattened so that all resampling iterations of all experiments can be executed in parallel.
future::plan("multisession")
+
+tasks = list(tsk("pima"), tsk("iris"))
+learner = lrn("classif.rpart")
+resampling = rsmp("cv", folds = 3)
+
+grid = benchmark_grid(tasks, learner, resampling)
+
+benchmark(grid)<BenchmarkResult> of 6 rows with 2 resampling runs
+ nr task_id learner_id resampling_id iters warnings errors
+ 1 pima classif.rpart cv 3 0 0
+ 2 iris classif.rpart cv 3 0 0The 2 experiments and the 3-fold cross-validation result in 6 jobs which are executed in parallel.
+The mlr3tuning package internally calls benchmark() during tuning. If the tuner is capable of suggesting multiple configurations per iteration (such as random search, grid search, or hyperband), these configurations represent individual experiments, and the loop flattening of benchmark() is triggered. E.g., all resampling iterations of all hyperparameter configurations on a grid can be executed in parallel.
future::plan("multisession")
+
+learner = lrn("classif.rpart")
+learner$param_set$values$cp = to_tune(0.001, 0.1)
+learner$param_set$values$minsplit = to_tune(1, 10)
+
+instance = tune(
+ tuner = tnr("random_search", batch_size = 5), # random search suggests 5 configurations per batch
+ task = tsk("pima"),
+ learner = learner,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ term_evals = 10
+)The batch size of 5 and the 3-fold cross-validation gives us 15 jobs. This is done twice because of the limit of 10 evaluations in total.
+Nested resampling results in two nested resampling loops. For this, an AutoTuner is passed to resample() or benchmark(). We can choose different parallelization backends for the inner and outer resampling loop, respectively. We just have to pass a list of backends.
# Runs the outer loop in parallel and the inner loop sequentially
+future::plan(list("multisession", "sequential"))
+
+# Runs the outer loop sequentially and the inner loop in parallel
+future::plan(list("sequential", "multisession"))
+
+learner = lrn("classif.rpart")
+learner$param_set$values$cp = to_tune(0.001, 0.1)
+learner$param_set$values$minsplit = to_tune(1, 10)
+
+rr = tune_nested(
+ tuner = tnr("random_search", batch_size = 5), # random search suggests 5 configurations per batch
+ task = tsk("pima"),
+ learner = learner,
+ inner_resampling = rsmp ("cv", folds = 3),
+ outer_resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ term_evals = 10
+)While nesting real parallelization backends is often unintended and causes unnecessary overhead, it is useful in some distributed computing setups. It can be achieved with future by forcing a fixed number of workers for each loop.
+# Runs both loops in parallel
+future::plan(list(future::tweak("multisession", workers = 2),
+ future::tweak("multisession", workers = 4)))This example would run on 8 cores (= 2 * 4) on the local machine.
The mlr3book includes a chapters on parallelization. The mlr3cheatsheets contain frequently used commands and workflows of mlr3.
+ + +This is the third part of the practical tuning series. The other parts can be found here:
+In this post, we implement a simple automated machine learning (AutoML) system which includes preprocessing, a switch between multiple learners and hyperparameter tuning. For this, we build a pipeline with the mlr3pipelines extension package. Additionally, we use nested resampling to get an unbiased performance estimate of our AutoML system.
+ +We load the mlr3verse package which pulls in the most important packages for this example.
+library(mlr3verse)We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented. The lgr package is used for logging in all mlr3 packages. The mlr3 logger prints the logging messages from the base package, whereas the bbotk logger is responsible for logging messages from the optimization packages (e.g. mlr3tuning ).
set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")In this example, we use the Pima Indians Diabetes data set which is used to to predict whether or not a patient has diabetes. The patients are characterized by 8 numeric features and some have missing values.
+task = tsk("pima")We use three popular machine learning algorithms: k-nearest-neighbors, support vector machines and random forests.
+learners = list(
+ lrn("classif.kknn", id = "kknn"),
+ lrn("classif.svm", id = "svm", type = "C-classification"),
+ lrn("classif.ranger", id = "ranger")
+)The PipeOpBranch allows us to specify multiple alternatives paths. In this graph, the paths lead to the different learner models. The selection hyperparameter controls which path is executed i.e., which learner is used to fit a model. It is important to use the PipeOpBranch after the branching so that the outputs are merged into one result object. We visualize the graph with branching below.
graph =
+ po("branch", options = c("kknn", "svm", "ranger")) %>>%
+ gunion(lapply(learners, po)) %>>%
+ po("unbranch")
+graph$plot(html = FALSE)
Alternatively, we can use the ppl()-shortcut to load a predefined graph from the mlr_graphs dictionary. For this, the learner list must be named.
learners = list(
+ kknn = lrn("classif.kknn", id = "kknn"),
+ svm = lrn("classif.svm", id = "svm", type = "C-classification"),
+ ranger = lrn("classif.ranger", id = "ranger")
+)
+
+graph = ppl("branch", lapply(learners, po))The task has missing data in five columns.
+round(task$missings() / task$nrow, 2)diabetes age glucose insulin mass pedigree pregnant pressure triceps
+ 0.00 0.00 0.01 0.49 0.01 0.00 0.00 0.05 0.30 The pipeline "robustify" function creates a preprocessing pipeline based on our task. The resulting pipeline imputes missing values with PipeOpImputeHist and creates a dummy column (PipeOpMissInd) which indicates the imputed missing values. Internally, this creates two paths and the results are combined with PipeOpFeatureUnion. In contrast to PipeOpBranch, both paths are executed. Additionally, "robustify" adds PipeOpEncode to encode factor columns and PipeOpRemoveConstants to remove features with a constant value.
graph = ppl("robustify", task = task, factors_to_numeric = TRUE) %>>%
+ graph
+plot(graph, html = FALSE)
We could also create the preprocessing pipeline manually.
+gunion(list(po("imputehist"),
+ po("missind", affect_columns = selector_type(c("numeric", "integer"))))) %>>%
+ po("featureunion") %>>%
+ po("encode") %>>%
+ po("removeconstants")Graph with 5 PipeOps:
+ ID State sccssors prdcssors
+ imputehist <<UNTRAINED>> featureunion
+ missind <<UNTRAINED>> featureunion
+ featureunion <<UNTRAINED>> encode imputehist,missind
+ encode <<UNTRAINED>> removeconstants featureunion
+ removeconstants <<UNTRAINED>> encodeWe use as_learner() to create a GraphLearner which encapsulates the pipeline and can be used like a learner.
graph_learner = as_learner(graph)The parameter set of the graph learner includes all hyperparameters from all contained learners. The hyperparameter ids are prefixed with the corresponding learner ids. The hyperparameter branch.selection controls which learner is used.
as.data.table(graph_learner$param_set)[, .(id, class, lower, upper, nlevels)] id class lower upper nlevels
+ 1: removeconstants_prerobustify.ratio ParamDbl 0 1 Inf
+ 2: removeconstants_prerobustify.rel_tol ParamDbl 0 Inf Inf
+ 3: removeconstants_prerobustify.abs_tol ParamDbl 0 Inf Inf
+ 4: removeconstants_prerobustify.na_ignore ParamLgl NA NA 2
+ 5: removeconstants_prerobustify.affect_columns ParamUty NA NA Inf
+ 6: imputehist.affect_columns ParamUty NA NA Inf
+ 7: missind.which ParamFct NA NA 2
+ 8: missind.type ParamFct NA NA 4
+ 9: missind.affect_columns ParamUty NA NA Inf
+10: imputesample.affect_columns ParamUty NA NA Inf
+11: encode.method ParamFct NA NA 5
+12: encode.affect_columns ParamUty NA NA Inf
+13: removeconstants_postrobustify.ratio ParamDbl 0 1 Inf
+14: removeconstants_postrobustify.rel_tol ParamDbl 0 Inf Inf
+15: removeconstants_postrobustify.abs_tol ParamDbl 0 Inf Inf
+16: removeconstants_postrobustify.na_ignore ParamLgl NA NA 2
+17: removeconstants_postrobustify.affect_columns ParamUty NA NA Inf
+18: kknn.k ParamInt 1 Inf Inf
+19: kknn.distance ParamDbl 0 Inf Inf
+20: kknn.kernel ParamFct NA NA 10
+21: kknn.scale ParamLgl NA NA 2
+22: kknn.ykernel ParamUty NA NA Inf
+23: kknn.store_model ParamLgl NA NA 2
+24: svm.cachesize ParamDbl -Inf Inf Inf
+25: svm.class.weights ParamUty NA NA Inf
+26: svm.coef0 ParamDbl -Inf Inf Inf
+27: svm.cost ParamDbl 0 Inf Inf
+28: svm.cross ParamInt 0 Inf Inf
+29: svm.decision.values ParamLgl NA NA 2
+30: svm.degree ParamInt 1 Inf Inf
+31: svm.epsilon ParamDbl 0 Inf Inf
+32: svm.fitted ParamLgl NA NA 2
+33: svm.gamma ParamDbl 0 Inf Inf
+34: svm.kernel ParamFct NA NA 4
+35: svm.nu ParamDbl -Inf Inf Inf
+36: svm.scale ParamUty NA NA Inf
+37: svm.shrinking ParamLgl NA NA 2
+38: svm.tolerance ParamDbl 0 Inf Inf
+39: svm.type ParamFct NA NA 2
+40: ranger.alpha ParamDbl -Inf Inf Inf
+41: ranger.always.split.variables ParamUty NA NA Inf
+42: ranger.class.weights ParamUty NA NA Inf
+43: ranger.holdout ParamLgl NA NA 2
+44: ranger.importance ParamFct NA NA 4
+45: ranger.keep.inbag ParamLgl NA NA 2
+46: ranger.max.depth ParamInt 0 Inf Inf
+47: ranger.min.node.size ParamInt 1 Inf Inf
+48: ranger.min.prop ParamDbl -Inf Inf Inf
+49: ranger.minprop ParamDbl -Inf Inf Inf
+50: ranger.mtry ParamInt 1 Inf Inf
+51: ranger.mtry.ratio ParamDbl 0 1 Inf
+52: ranger.num.random.splits ParamInt 1 Inf Inf
+53: ranger.num.threads ParamInt 1 Inf Inf
+54: ranger.num.trees ParamInt 1 Inf Inf
+55: ranger.oob.error ParamLgl NA NA 2
+56: ranger.regularization.factor ParamUty NA NA Inf
+57: ranger.regularization.usedepth ParamLgl NA NA 2
+58: ranger.replace ParamLgl NA NA 2
+59: ranger.respect.unordered.factors ParamFct NA NA 3
+60: ranger.sample.fraction ParamDbl 0 1 Inf
+61: ranger.save.memory ParamLgl NA NA 2
+62: ranger.scale.permutation.importance ParamLgl NA NA 2
+63: ranger.se.method ParamFct NA NA 2
+64: ranger.seed ParamInt -Inf Inf Inf
+65: ranger.split.select.weights ParamUty NA NA Inf
+66: ranger.splitrule ParamFct NA NA 3
+67: ranger.verbose ParamLgl NA NA 2
+68: ranger.write.forest ParamLgl NA NA 2
+69: branch.selection ParamFct NA NA 3
+ id class lower upper nlevelsWe will only tune one hyperparameter for each learner in this example. Additionally, we tune the branching parameter which selects one of the three learners. We have to specify that a hyperparameter is only valid for a certain learner by using depends = branch.selection == <learner_id>.
# branch
+graph_learner$param_set$values$branch.selection =
+ to_tune(c("kknn", "svm", "ranger"))
+
+# kknn
+graph_learner$param_set$values$kknn.k =
+ to_tune(p_int(3, 50, logscale = TRUE, depends = branch.selection == "kknn"))
+
+# svm
+graph_learner$param_set$values$svm.cost =
+ to_tune(p_dbl(-1, 1, trafo = function(x) 10^x, depends = branch.selection == "svm"))
+
+# ranger
+graph_learner$param_set$values$ranger.mtry =
+ to_tune(p_int(1, 8, depends = branch.selection == "ranger"))
+
+# short learner id for printing
+graph_learner$id = "graph_learner"We define a tuning instance and select a random search which is stopped after 20 evaluated configurations.
+instance = tune(
+ tuner = tnr("random_search"),
+ task = task,
+ learner = graph_learner,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ term_evals = 20
+)The following shows a quick way to visualize the tuning results.
+autoplot(instance, type = "marginal",
+ cols_x = c("x_domain_kknn.k", "x_domain_svm.cost", "ranger.mtry"))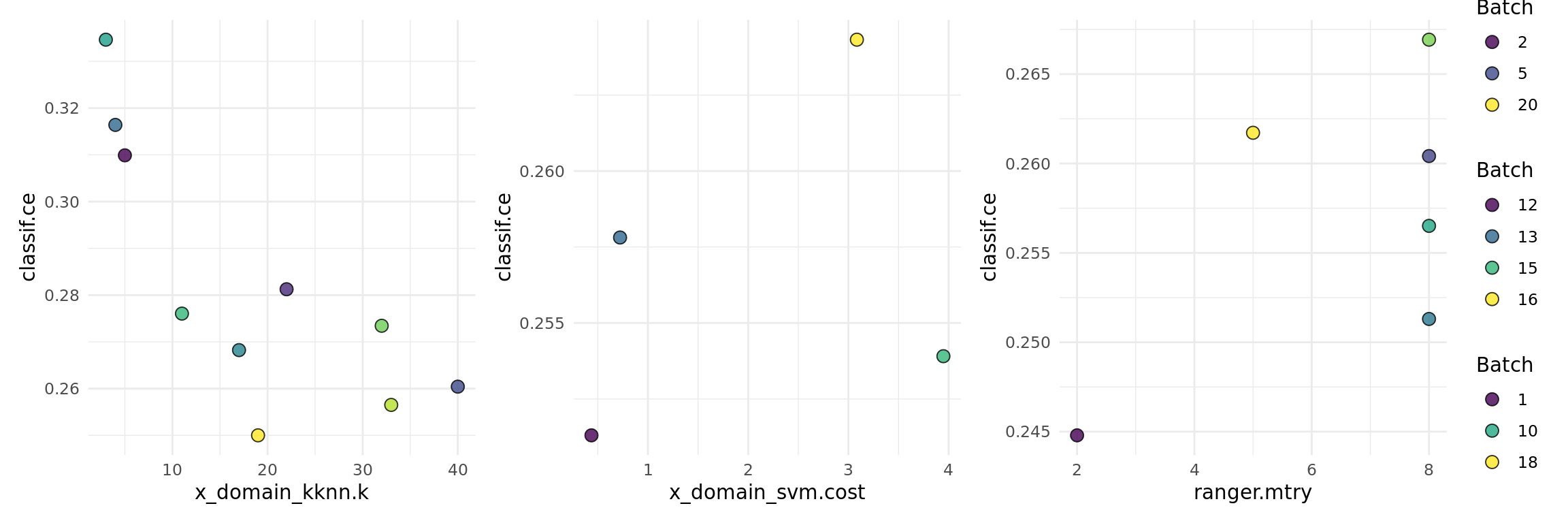
We add the optimized hyperparameters to the graph learner and train the learner on the full dataset.
+learner = as_learner(graph)
+learner$param_set$values = instance$result_learner_param_vals
+learner$train(task)The trained model can now be used to make predictions on new data. A common mistake is to report the performance estimated on the resampling sets on which the tuning was performed (instance$result_y) as the model’s performance. Instead we have to use nested resampling to get an unbiased performance estimate.
We use nested resampling to get an unbiased estimate of the predictive performance of our graph learner.
+graph_learner = as_learner(graph)
+graph_learner$param_set$values$branch.selection =
+ to_tune(c("kknn", "svm", "ranger"))
+graph_learner$param_set$values$kknn.k =
+ to_tune(p_int(3, 50, logscale = TRUE, depends = branch.selection == "kknn"))
+graph_learner$param_set$values$svm.cost =
+ to_tune(p_dbl(-1, 1, trafo = function(x) 10^x, depends = branch.selection == "svm"))
+graph_learner$param_set$values$ranger.mtry =
+ to_tune(p_int(1, 8, depends = branch.selection == "ranger"))
+graph_learner$id = "graph_learner"
+
+inner_resampling = rsmp("cv", folds = 3)
+at = AutoTuner$new(
+ learner = graph_learner,
+ resampling = inner_resampling,
+ measure = msr("classif.ce"),
+ terminator = trm("evals", n_evals = 10),
+ tuner = tnr("random_search")
+)
+
+outer_resampling = rsmp("cv", folds = 3)
+rr = resample(task, at, outer_resampling, store_models = TRUE)We check the inner tuning results for stable hyperparameters. This means that the selected hyperparameters should not vary too much. We might observe unstable models in this example because the small data set and the low number of resampling iterations might introduce too much randomness. Usually, we aim for the selection of stable hyperparameters for all outer training sets.
+extract_inner_tuning_results(rr)Next, we want to compare the predictive performances estimated on the outer resampling to the inner resampling. Significantly lower predictive performances on the outer resampling indicate that the models with the optimized hyperparameters overfit the data.
+rr$score()[, .(iteration, task_id, learner_id, resampling_id, classif.ce)] iteration task_id learner_id resampling_id classif.ce
+1: 1 pima graph_learner.tuned cv 0.2265625
+2: 2 pima graph_learner.tuned cv 0.2382812
+3: 3 pima graph_learner.tuned cv 0.2656250The aggregated performance of all outer resampling iterations is essentially the unbiased performance of the graph learner with optimal hyperparameter found by random search.
+rr$aggregate()classif.ce
+ 0.2434896 Applying nested resampling can be shortened by using the tune_nested()-shortcut.
graph_learner = as_learner(graph)
+graph_learner$param_set$values$branch.selection =
+ to_tune(c("kknn", "svm", "ranger"))
+graph_learner$param_set$values$kknn.k =
+ to_tune(p_int(3, 50, logscale = TRUE, depends = branch.selection == "kknn"))
+graph_learner$param_set$values$svm.cost =
+ to_tune(p_dbl(-1, 1, trafo = function(x) 10^x, depends = branch.selection == "svm"))
+graph_learner$param_set$values$ranger.mtry =
+ to_tune(p_int(1, 8, depends = branch.selection == "ranger"))
+graph_learner$id = "graph_learner"
+
+rr = tune_nested(
+ tuner = tnr("random_search"),
+ task = task,
+ learner = graph_learner,
+ inner_resampling = rsmp("cv", folds = 3),
+ outer_resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+ term_evals = 10,
+)The mlr3book includes chapters on pipelines and hyperparameter tuning. The mlr3cheatsheets contain frequently used commands and workflows of mlr3.
+ + +This is the second part of the practical tuning series. The other parts can be found here:
+In this post, we build a simple preprocessing pipeline and tune it. For this, we are using the mlr3pipelines extension package. First, we start by imputing missing values in the Pima Indians Diabetes data set. After that, we encode a factor column to numerical dummy columns in the data set. Next, we combine both preprocessing steps to a Graph and create a GraphLearner. Finally, nested resampling is used to compare the performance of two imputation methods.
We load the mlr3verse package which pulls in the most important packages for this example.
+library(mlr3verse)We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented. The lgr package is used for logging in all mlr3 packages. The mlr3 logger prints the logging messages from the base package, whereas the bbotk logger is responsible for logging messages from the optimization packages (e.g. mlr3tuning ).
set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")In this example, we use the Pima Indians Diabetes data set which is used to predict whether or not a patient has diabetes. The patients are characterized by 8 numeric features of which some have missing values. We alter the data set by categorizing the feature pressure (blood pressure) into the categories "low", "mid", and "high".
# retrieve the task from mlr3
+task = tsk("pima")
+
+# create data frame with categorized pressure feature
+data = task$data(cols = "pressure")
+breaks = quantile(data$pressure, probs = c(0, 0.33, 0.66, 1), na.rm = TRUE)
+data$pressure = cut(data$pressure, breaks, labels = c("low", "mid", "high"))
+
+# overwrite the feature in the task
+task$cbind(data)
+
+# generate a quick textual overview
+skimr::skim(task$data())| Name | +task$data() | +
| Number of rows | +768 | +
| Number of columns | +9 | +
| Key | +NULL | +
| _______________________ | ++ |
| Column type frequency: | ++ |
| factor | +2 | +
| numeric | +7 | +
| ________________________ | ++ |
| Group variables | +None | +
Variable type: factor
+| skim_variable | +n_missing | +complete_rate | +ordered | +n_unique | +top_counts | +
|---|---|---|---|---|---|
| diabetes | +0 | +1.00 | +FALSE | +2 | +neg: 500, pos: 268 | +
| pressure | +36 | +0.95 | +FALSE | +3 | +low: 282, mid: 245, hig: 205 | +
Variable type: numeric
+| skim_variable | +n_missing | +complete_rate | +mean | +sd | +p0 | +p25 | +p50 | +p75 | +p100 | +hist | +
|---|---|---|---|---|---|---|---|---|---|---|
| age | +0 | +1.00 | +33.24 | +11.76 | +21.00 | +24.00 | +29.00 | +41.00 | +81.00 | +▇▃▁▁▁ | +
| glucose | +5 | +0.99 | +121.69 | +30.54 | +44.00 | +99.00 | +117.00 | +141.00 | +199.00 | +▁▇▇▃▂ | +
| insulin | +374 | +0.51 | +155.55 | +118.78 | +14.00 | +76.25 | +125.00 | +190.00 | +846.00 | +▇▂▁▁▁ | +
| mass | +11 | +0.99 | +32.46 | +6.92 | +18.20 | +27.50 | +32.30 | +36.60 | +67.10 | +▅▇▃▁▁ | +
| pedigree | +0 | +1.00 | +0.47 | +0.33 | +0.08 | +0.24 | +0.37 | +0.63 | +2.42 | +▇▃▁▁▁ | +
| pregnant | +0 | +1.00 | +3.85 | +3.37 | +0.00 | +1.00 | +3.00 | +6.00 | +17.00 | +▇▃▂▁▁ | +
| triceps | +227 | +0.70 | +29.15 | +10.48 | +7.00 | +22.00 | +29.00 | +36.00 | +99.00 | +▆▇▁▁▁ | +
We choose the xgboost algorithm from the xgboost package as learner.
+learner = lrn("classif.xgboost", nrounds = 100, id = "xgboost", verbose = 0)The task has missing data in five columns.
+round(task$missings() / task$nrow, 2)diabetes age glucose insulin mass pedigree pregnant pressure triceps
+ 0.00 0.00 0.01 0.49 0.01 0.00 0.00 0.05 0.30 The xgboost learner has an internal method for handling missing data but some learners cannot handle missing values. We will try to beat the internal method in terms of predictive performance. The mlr3pipelines package offers various methods to impute missing values.
mlr_pipeops$keys("^impute")[1] "imputeconstant" "imputehist" "imputelearner" "imputemean" "imputemedian" "imputemode"
+[7] "imputeoor" "imputesample" We choose the PipeOpImputeOOR that adds the new factor level ".MISSING". to factorial features and imputes numerical features by constant values shifted below the minimum (default) or above the maximum.
imputer = po("imputeoor")
+print(imputer)PipeOp: <imputeoor> (not trained)
+values: <min=TRUE, offset=1, multiplier=1>
+Input channels <name [train type, predict type]>:
+ input [Task,Task]
+Output channels <name [train type, predict type]>:
+ output [Task,Task]As the output suggests, the in- and output of this pipe operator is a Task for both the training and the predict step. We can manually train the pipe operator to check its functionality:
task_imputed = imputer$train(list(task))[[1]]
+task_imputed$missings()diabetes age pedigree pregnant glucose insulin mass pressure triceps
+ 0 0 0 0 0 0 0 0 0 Let’s compare an observation with missing values to the observation with imputed observation.
+rbind(
+ task$data()[8,],
+ task_imputed$data()[8,]
+) diabetes age glucose insulin mass pedigree pregnant pressure triceps
+1: neg 29 115 NA 35.3 0.134 10 <NA> NA
+2: neg 29 115 -819 35.3 0.134 10 .MISSING -86Note that OOR imputation is in particular useful for tree-based models, but should not be used for linear models or distance-based models.
+The xgboost learner cannot handle categorical features. Therefore, we must to convert factor columns to numerical dummy columns. For this, we argument the xgboost learner with automatic factor encoding.
The PipeOpEncode encodes factor columns with one of six methods. In this example, we use one-hot encoding which creates a new binary column for each factor level.
factor_encoding = po("encode", method = "one-hot")We manually trigger the encoding on the task.
+factor_encoding$train(list(task))$output
+<TaskClassif:pima> (768 x 11): Pima Indian Diabetes
+* Target: diabetes
+* Properties: twoclass
+* Features (10):
+ - dbl (10): age, glucose, insulin, mass, pedigree, pregnant, pressure.high, pressure.low, pressure.mid,
+ tricepsThe factor column pressure has been converted to the three binary columns "pressure.low", "pressure.mid", and "pressure.high".
We created two preprocessing steps which could be used to create a new task with encoded factor variables and imputed missing values. However, if we do this before resampling, information from the test can leak into our training step which typically leads to overoptimistic performance measures. To avoid this, we add the preprocessing steps to the Learner itself, creating a GraphLearner. For this, we create a Graph first.
graph = po("encode") %>>%
+ po("imputeoor") %>>%
+ learner
+plot(graph, html = FALSE)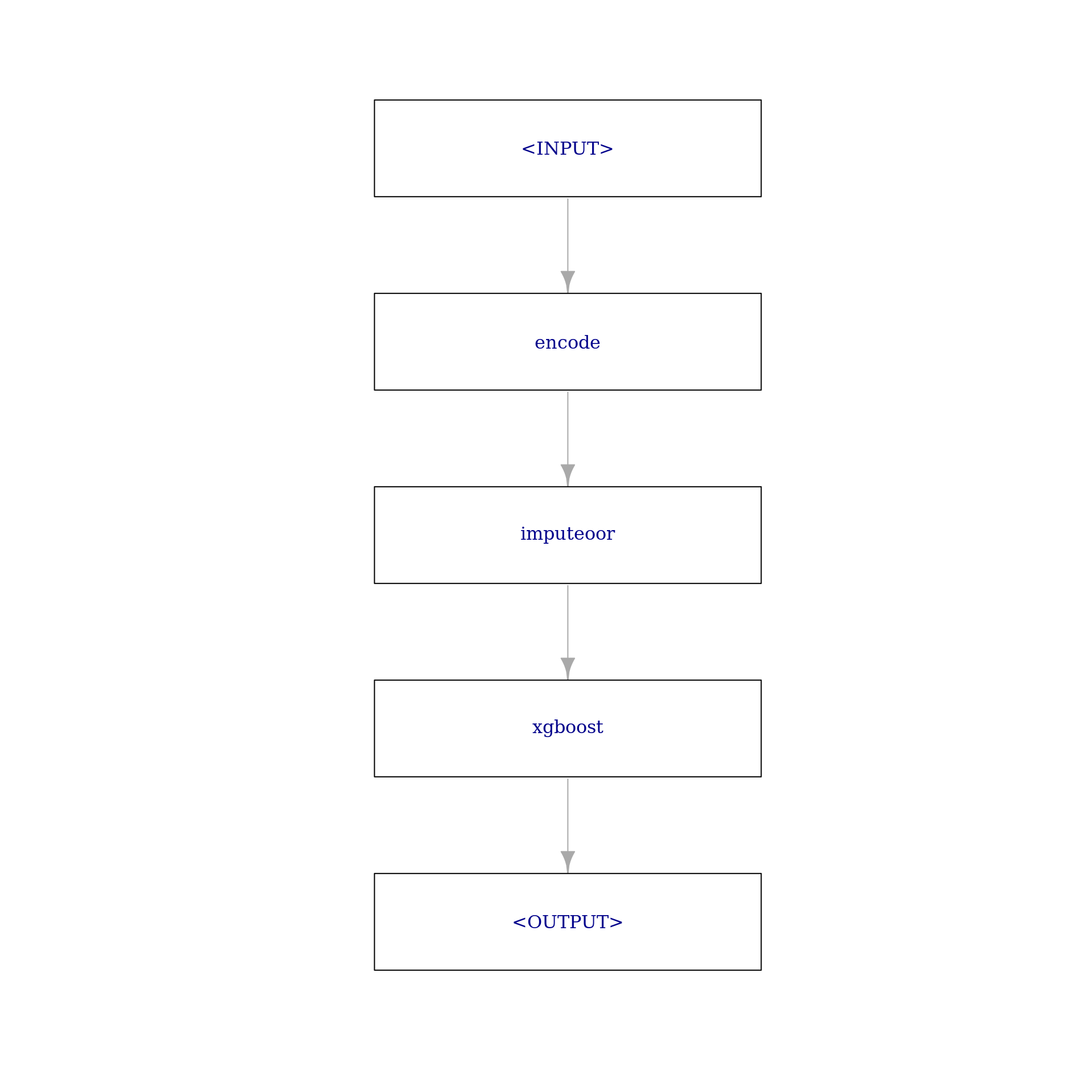
We use as_learner() to wrap the Graph into a GraphLearner with which allows us to use the graph like a normal learner.
graph_learner = as_learner(graph)
+
+# short learner id for printing
+graph_learner$id = "graph_learner"The GraphLearner can be trained and used for making predictions. Instead of calling $train() or $predict() manually, we will directly use it for resampling. We choose a 3-fold cross-validation as the resampling strategy.
resampling = rsmp("cv", folds = 3)
+
+rr = resample(task = task, learner = graph_learner, resampling = resampling)rr$score()[, c("iteration", "task_id", "learner_id", "resampling_id", "classif.ce"), with = FALSE] iteration task_id learner_id resampling_id classif.ce
+1: 1 pima graph_learner cv 0.2851562
+2: 2 pima graph_learner cv 0.2460938
+3: 3 pima graph_learner cv 0.2968750For each resampling iteration, the following steps are performed:
+Next is the predict step:
+By following this procedure, it is guaranteed that no information can leak from the training step to the predict step.
+Let’s have a look at the parameter set of the GraphLearner. It consists of the xgboost hyperparameters, and additionally, the parameter of the PipeOp encode and imputeoor. All hyperparameters are prefixed with the id of the respective PipeOp or learner.
as.data.table(graph_learner$param_set)[, c("id", "class", "lower", "upper", "nlevels"), with = FALSE] id class lower upper nlevels
+ 1: encode.method ParamFct NA NA 5
+ 2: encode.affect_columns ParamUty NA NA Inf
+ 3: imputeoor.min ParamLgl NA NA 2
+ 4: imputeoor.offset ParamDbl 0 Inf Inf
+ 5: imputeoor.multiplier ParamDbl 0 Inf Inf
+ 6: imputeoor.affect_columns ParamUty NA NA Inf
+ 7: xgboost.alpha ParamDbl 0 Inf Inf
+ 8: xgboost.approxcontrib ParamLgl NA NA 2
+ 9: xgboost.base_score ParamDbl -Inf Inf Inf
+10: xgboost.booster ParamFct NA NA 3
+11: xgboost.callbacks ParamUty NA NA Inf
+12: xgboost.colsample_bylevel ParamDbl 0 1 Inf
+13: xgboost.colsample_bynode ParamDbl 0 1 Inf
+14: xgboost.colsample_bytree ParamDbl 0 1 Inf
+15: xgboost.disable_default_eval_metric ParamLgl NA NA 2
+16: xgboost.early_stopping_rounds ParamInt 1 Inf Inf
+17: xgboost.early_stopping_set ParamFct NA NA 3
+18: xgboost.eta ParamDbl 0 1 Inf
+19: xgboost.eval_metric ParamUty NA NA Inf
+20: xgboost.feature_selector ParamFct NA NA 5
+21: xgboost.feval ParamUty NA NA Inf
+22: xgboost.gamma ParamDbl 0 Inf Inf
+23: xgboost.grow_policy ParamFct NA NA 2
+24: xgboost.interaction_constraints ParamUty NA NA Inf
+25: xgboost.iterationrange ParamUty NA NA Inf
+26: xgboost.lambda ParamDbl 0 Inf Inf
+27: xgboost.lambda_bias ParamDbl 0 Inf Inf
+28: xgboost.max_bin ParamInt 2 Inf Inf
+29: xgboost.max_delta_step ParamDbl 0 Inf Inf
+30: xgboost.max_depth ParamInt 0 Inf Inf
+31: xgboost.max_leaves ParamInt 0 Inf Inf
+32: xgboost.maximize ParamLgl NA NA 2
+33: xgboost.min_child_weight ParamDbl 0 Inf Inf
+34: xgboost.missing ParamDbl -Inf Inf Inf
+35: xgboost.monotone_constraints ParamUty NA NA Inf
+36: xgboost.normalize_type ParamFct NA NA 2
+37: xgboost.nrounds ParamInt 1 Inf Inf
+38: xgboost.nthread ParamInt 1 Inf Inf
+39: xgboost.ntreelimit ParamInt 1 Inf Inf
+40: xgboost.num_parallel_tree ParamInt 1 Inf Inf
+41: xgboost.objective ParamUty NA NA Inf
+42: xgboost.one_drop ParamLgl NA NA 2
+43: xgboost.outputmargin ParamLgl NA NA 2
+44: xgboost.predcontrib ParamLgl NA NA 2
+45: xgboost.predictor ParamFct NA NA 2
+46: xgboost.predinteraction ParamLgl NA NA 2
+47: xgboost.predleaf ParamLgl NA NA 2
+48: xgboost.print_every_n ParamInt 1 Inf Inf
+49: xgboost.process_type ParamFct NA NA 2
+50: xgboost.rate_drop ParamDbl 0 1 Inf
+51: xgboost.refresh_leaf ParamLgl NA NA 2
+52: xgboost.reshape ParamLgl NA NA 2
+53: xgboost.seed_per_iteration ParamLgl NA NA 2
+54: xgboost.sampling_method ParamFct NA NA 2
+55: xgboost.sample_type ParamFct NA NA 2
+56: xgboost.save_name ParamUty NA NA Inf
+57: xgboost.save_period ParamInt 0 Inf Inf
+58: xgboost.scale_pos_weight ParamDbl -Inf Inf Inf
+59: xgboost.skip_drop ParamDbl 0 1 Inf
+60: xgboost.strict_shape ParamLgl NA NA 2
+61: xgboost.subsample ParamDbl 0 1 Inf
+62: xgboost.top_k ParamInt 0 Inf Inf
+63: xgboost.training ParamLgl NA NA 2
+64: xgboost.tree_method ParamFct NA NA 5
+65: xgboost.tweedie_variance_power ParamDbl 1 2 Inf
+66: xgboost.updater ParamUty NA NA Inf
+67: xgboost.verbose ParamInt 0 2 3
+68: xgboost.watchlist ParamUty NA NA Inf
+69: xgboost.xgb_model ParamUty NA NA Inf
+ id class lower upper nlevelsWe will tune the encode method.
+graph_learner$param_set$values$encode.method = to_tune(c("one-hot", "treatment"))We define a tuning instance and use grid search since we want to try all encode methods.
+instance = tune(
+ tuner = tnr("grid_search"),
+ task = task,
+ learner = graph_learner,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce")
+)The archive shows us the performance of the model with different encoding methods.
+print(instance$archive)<ArchiveTuning> with 2 evaluations
+ encode.method classif.ce batch_nr warnings errors
+1: treatment 0.26 1 0 0
+2: one-hot 0.26 2 0 0We create one GraphLearner with imputeoor and test it against a GraphLearner that uses the internal imputation method of xgboost. Applying nested resampling ensures a fair comparison of the predictive performances.
graph_1 = po("encode") %>>%
+ learner
+graph_learner_1 = GraphLearner$new(graph_1)
+
+graph_learner_1$param_set$values$encode.method = to_tune(c("one-hot", "treatment"))
+
+at_1 = AutoTuner$new(
+ learner = graph_learner_1,
+ resampling = resampling,
+ measure = msr("classif.ce"),
+ terminator = trm("none"),
+ tuner = tnr("grid_search"),
+ store_models = TRUE
+)graph_2 = po("encode") %>>%
+ po("imputeoor") %>>%
+ learner
+graph_learner_2 = GraphLearner$new(graph_2)
+
+graph_learner_2$param_set$values$encode.method = to_tune(c("one-hot", "treatment"))
+
+at_2 = AutoTuner$new(
+ learner = graph_learner_2,
+ resampling = resampling,
+ measure = msr("classif.ce"),
+ terminator = trm("none"),
+ tuner = tnr("grid_search"),
+ store_models = TRUE
+)We run the benchmark.
+resampling_outer = rsmp("cv", folds = 3)
+design = benchmark_grid(task, list(at_1, at_2), resampling_outer)
+
+bmr = benchmark(design, store_models = TRUE)We compare the aggregated performances on the outer test sets which give us an unbiased performance estimate of the GraphLearners with the different encoding methods.
bmr$aggregate() nr task_id learner_id resampling_id iters classif.ce
+1: 1 pima encode.xgboost.tuned cv 3 0.2578125
+2: 2 pima encode.imputeoor.xgboost.tuned cv 3 0.2630208
+Hidden columns: resample_resultautoplot(bmr)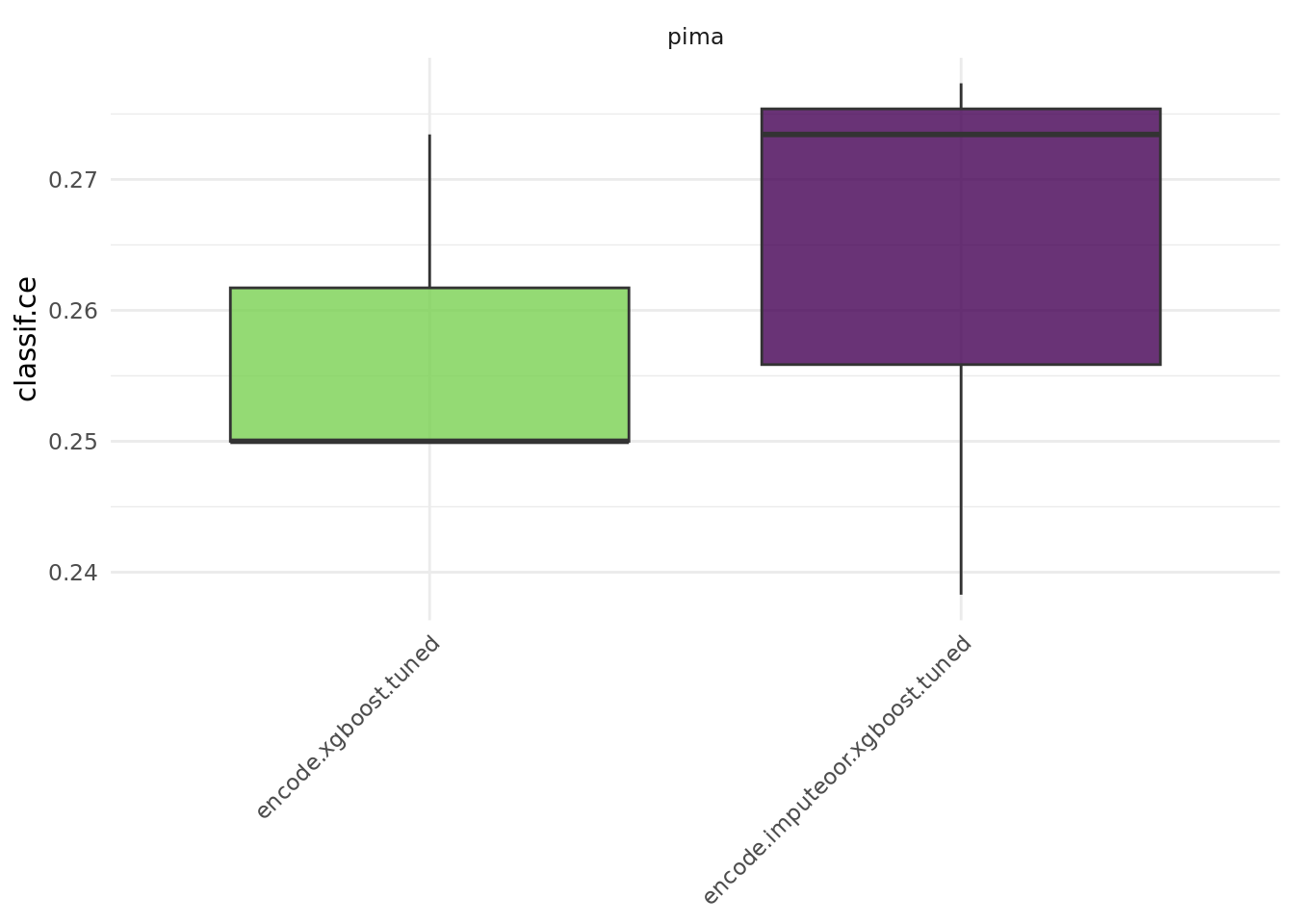
Note that in practice, it is required to tune preprocessing hyperparameters jointly with the hyperparameters of the learner. Otherwise, comparing preprocessing steps is not feasible and can lead to wrong conclusions.
+Applying nested resampling can be shortened by using the auto_tuner()-shortcut.
graph_1 = po("encode") %>>% learner
+graph_learner_1 = as_learner(graph_1)
+graph_learner_1$param_set$values$encode.method = to_tune(c("one-hot", "treatment"))
+
+at_1 = auto_tuner(
+ method = "grid_search",
+ learner = graph_learner_1,
+ resampling = resampling,
+ measure = msr("classif.ce"),
+ store_models = TRUE)
+
+graph_2 = po("encode") %>>% po("imputeoor") %>>% learner
+graph_learner_2 = as_learner(graph_2)
+graph_learner_2$param_set$values$encode.method = to_tune(c("one-hot", "treatment"))
+
+at_2 = auto_tuner(
+ method = "grid_search",
+ learner = graph_learner_2,
+ resampling = resampling,
+ measure = msr("classif.ce"),
+ store_models = TRUE)
+
+design = benchmark_grid(task, list(at_1, at_2), rsmp("cv", folds = 3))
+
+bmr = benchmark(design, store_models = TRUE)We train the chosen GraphLearner with the AutoTuner to get a final model with optimized hyperparameters.
at_2$train(task)The trained model can now be used to make predictions on new data at_2$predict(). The pipeline ensures that the preprocessing is always a part of the train and predict step.
The mlr3book includes chapters on pipelines and hyperparameter tuning. The mlr3cheatsheets contain frequently used commands and workflows of mlr3.
+ + +requireNamespace("DiceKriging")Loading required namespace: DiceKrigingThis is the first part of the practical tuning series. The other parts can be found here:
+In this post, we demonstrate how to optimize the hyperparameters of a support vector machine (SVM). We are using the mlr3 machine learning framework with the mlr3tuning extension package.
+First, we start by showing the basic building blocks of mlr3tuning and tune the cost and gamma hyperparameters of an SVM with a radial basis function on the Iris data set. After that, we use transformations to tune the both hyperparameters on the logarithmic scale. Next, we explain the importance of dependencies to tune hyperparameters like degree which are dependent on the choice of kernel. After that, we fit an SVM with optimized hyperparameters on the full dataset. Finally, nested resampling is used to compute an unbiased performance estimate of our tuned SVM.
We load the mlr3verse package which pulls in the most important packages for this example.
+library(mlr3verse)We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented. The lgr package is used for logging in all mlr3 packages. The mlr3 logger prints the logging messages from the base package, whereas the bbotk logger is responsible for logging messages from the optimization packages (e.g. mlr3tuning ).
set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")In the example, we use the Iris data set which classifies 150 flowers in three species of Iris. The flowers are characterized by sepal length and width and petal length and width. The Iris data set allows us to quickly fit models to it. However, the influence of hyperparameter tuning on the predictive performance might be minor. Other data sets might give more meaningful tuning results.
+# retrieve the task from mlr3
+task = tsk("iris")
+
+# generate a quick textual overview using the skimr package
+skimr::skim(task$data())| Name | +task$data() | +
| Number of rows | +150 | +
| Number of columns | +5 | +
| Key | +NULL | +
| _______________________ | ++ |
| Column type frequency: | ++ |
| factor | +1 | +
| numeric | +4 | +
| ________________________ | ++ |
| Group variables | +None | +
Variable type: factor
+| skim_variable | +n_missing | +complete_rate | +ordered | +n_unique | +top_counts | +
|---|---|---|---|---|---|
| Species | +0 | +1 | +FALSE | +3 | +set: 50, ver: 50, vir: 50 | +
Variable type: numeric
+| skim_variable | +n_missing | +complete_rate | +mean | +sd | +p0 | +p25 | +p50 | +p75 | +p100 | +hist | +
|---|---|---|---|---|---|---|---|---|---|---|
| Petal.Length | +0 | +1 | +3.76 | +1.77 | +1.0 | +1.6 | +4.35 | +5.1 | +6.9 | +▇▁▆▇▂ | +
| Petal.Width | +0 | +1 | +1.20 | +0.76 | +0.1 | +0.3 | +1.30 | +1.8 | +2.5 | +▇▁▇▅▃ | +
| Sepal.Length | +0 | +1 | +5.84 | +0.83 | +4.3 | +5.1 | +5.80 | +6.4 | +7.9 | +▆▇▇▅▂ | +
| Sepal.Width | +0 | +1 | +3.06 | +0.44 | +2.0 | +2.8 | +3.00 | +3.3 | +4.4 | +▁▆▇▂▁ | +
We choose the support vector machine implementation from the e1071 package (which is based on LIBSVM) and use it as a classification machine by setting type to "C-classification".
learner = lrn("classif.svm", type = "C-classification", kernel = "radial")For tuning, it is important to create a search space that defines the type and range of the hyperparameters. A learner stores all information about its hyperparameters in the slot $param_set. Not all parameters are tunable. We have to choose a subset of the hyperparameters we want to tune.
as.data.table(learner$param_set)[, .(id, class, lower, upper, nlevels)]We use the to_tune() function to define the range over which the hyperparameter should be tuned. We opt for the cost and gamma hyperparameters of the radial kernel and set the tuning ranges with lower and upper bounds.
learner$param_set$values$cost = to_tune(0.1, 10)
+learner$param_set$values$gamma = to_tune(0, 5)We specify how to evaluate the performance of the different hyperparameter configurations. For this, we choose 3-fold cross validation as the resampling strategy and the classification error as the performance measure.
+resampling = rsmp("cv", folds = 3)
+measure = msr("classif.ce")Usually, we have to select a budget for the tuning. This is done by choosing a Terminator, which stops the tuning e.g. after a performance level is reached or after a given time. However, some tuners like grid search terminate themselves. In this case, we choose a terminator that never stops and the tuning is not stopped before all grid points are evaluated.
terminator = trm("none")At this point, we can construct a TuningInstanceSingleCrit that describes the tuning problem.
instance = TuningInstanceSingleCrit$new(
+ task = task,
+ learner = learner,
+ resampling = resampling,
+ measure = measure,
+ terminator = terminator
+)
+
+print(instance)<TuningInstanceSingleCrit>
+* State: Not optimized
+* Objective: <ObjectiveTuning:classif.svm_on_iris>
+* Search Space:
+ id class lower upper nlevels
+1: cost ParamDbl 0.1 10 Inf
+2: gamma ParamDbl 0.0 5 Inf
+* Terminator: <TerminatorNone>Finally, we have to choose a Tuner. Grid Search discretizes numeric parameters into a given resolution and constructs a grid from the Cartesian product of these sets. Categorical parameters produce a grid over all levels specified in the search space. In this example, we only use a resolution of 5 to keep the runtime low. Usually, a higher resolution is used to create a denser grid.
tuner = tnr("grid_search", resolution = 5)
+
+print(tuner)<TunerGridSearch>: Grid Search
+* Parameters: resolution=5, batch_size=1
+* Parameter classes: ParamLgl, ParamInt, ParamDbl, ParamFct
+* Properties: dependencies, single-crit, multi-crit
+* Packages: mlr3tuningWe can preview the proposed configurations by using generate_design_grid(). This function is internally executed by TunerGridSearch.
generate_design_grid(learner$param_set$search_space(), resolution = 5)<Design> with 25 rows:
+ cost gamma
+ 1: 0.100 0.00
+ 2: 0.100 1.25
+ 3: 0.100 2.50
+ 4: 0.100 3.75
+ 5: 0.100 5.00
+ 6: 2.575 0.00
+ 7: 2.575 1.25
+ 8: 2.575 2.50
+ 9: 2.575 3.75
+10: 2.575 5.00
+11: 5.050 0.00
+12: 5.050 1.25
+13: 5.050 2.50
+14: 5.050 3.75
+15: 5.050 5.00
+16: 7.525 0.00
+17: 7.525 1.25
+18: 7.525 2.50
+19: 7.525 3.75
+20: 7.525 5.00
+21: 10.000 0.00
+22: 10.000 1.25
+23: 10.000 2.50
+24: 10.000 3.75
+25: 10.000 5.00
+ cost gammaWe trigger the tuning by passing the TuningInstanceSingleCrit to the $optimize() method of the Tuner. The instance is modified in-place.
tuner$optimize(instance) cost gamma learner_param_vals x_domain classif.ce
+1: 5.05 1.25 <list[4]> <list[2]> 0.06We plot the performances depending on the evaluated cost and gamma values.
autoplot(instance, type = "surface", cols_x = c("cost", "gamma"),
+ learner = lrn("regr.km"))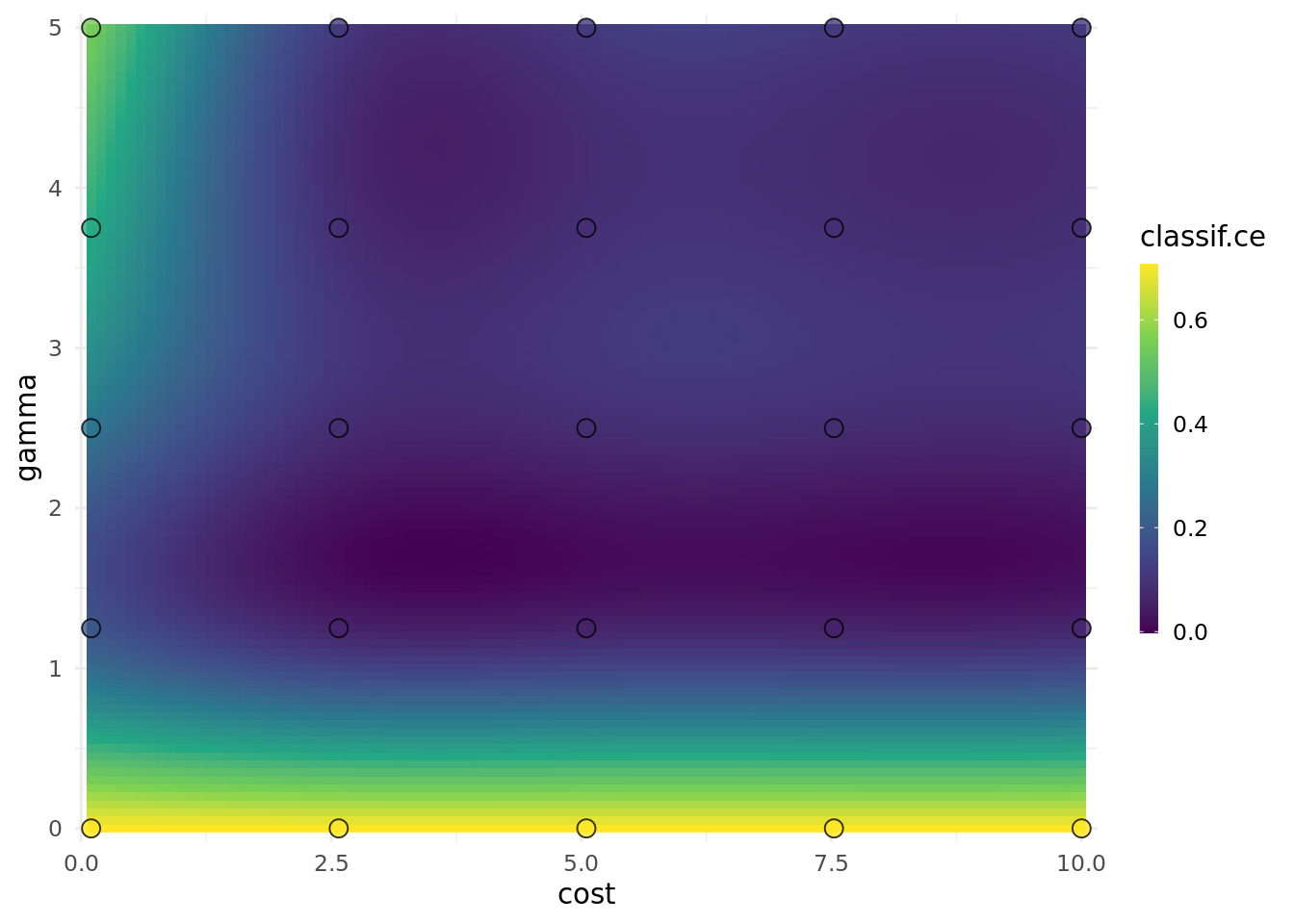
The points mark the evaluated cost and gamma values. We should not infer the performance of new values from the heatmap since it is only an interpolation. However, we can see the general interaction between the hyperparameters.
Tuning a learner can be shortened by using the tune()-shortcut.
learner = lrn("classif.svm", type = "C-classification", kernel = "radial")
+learner$param_set$values$cost = to_tune(0.1, 10)
+learner$param_set$values$gamma = to_tune(0, 5)
+
+instance = tune(
+ tuner = tnr("grid_search", resolution = 5),
+ task = tsk("iris"),
+ learner = learner,
+ resampling = rsmp ("holdout"),
+ measure = msr("classif.ce")
+)Next, we want to tune the cost and gamma hyperparameter more efficiently. It is recommended to tune cost and gamma on the logarithmic scale (Hsu, Chang, and Lin 2003). The log transformation emphasizes smaller cost and gamma values but also creates large values. Therefore, we use a log transformation to emphasize this region of the search space with a denser grid.
Generally speaking, transformations can be used to convert hyperparameters to a new scale. These transformations are applied before the proposed configuration is passed to the Learner. We can directly define the transformation in the to_tune() function. The lower and upper bound is set on the original scale.
learner = lrn("classif.svm", type = "C-classification", kernel = "radial")
+
+# tune from 2^-15 to 2^15 on a log scale
+learner$param_set$values$cost = to_tune(p_dbl(-15, 15, trafo = function(x) 2^x))
+
+# tune from 2^-15 to 2^5 on a log scale
+learner$param_set$values$gamma = to_tune(p_dbl(-15, 5, trafo = function(x) 2^x))Transformations to the log scale are the ones most commonly used. We can use a shortcut for this transformation. The lower and upper bound is set on the transformed scale.
+learner$param_set$values$cost = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$gamma = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))We use the tune()-shortcut to run the tuning.
instance = tune(
+ tuner = tnr("grid_search", resolution = 5),
+ task = task,
+ learner = learner,
+ resampling = resampling,
+ measure = measure
+)The hyperparameter values after the transformation are stored in the x_domain column as lists. We can expand these lists into multiple columns by using as.data.table(). The hyperparameter names are prefixed by x_domain.
as.data.table(instance$archive)[, .(cost, gamma, x_domain_cost, x_domain_gamma)] cost gamma x_domain_cost x_domain_gamma
+ 1: -5.756463 -11.512925 3.162278e-03 1.000000e-05
+ 2: 0.000000 11.512925 1.000000e+00 1.000000e+05
+ 3: -11.512925 11.512925 1.000000e-05 1.000000e+05
+ 4: 11.512925 5.756463 1.000000e+05 3.162278e+02
+ 5: -11.512925 0.000000 1.000000e-05 1.000000e+00
+ 6: 11.512925 -5.756463 1.000000e+05 3.162278e-03
+ 7: 11.512925 0.000000 1.000000e+05 1.000000e+00
+ 8: -11.512925 5.756463 1.000000e-05 3.162278e+02
+ 9: 0.000000 0.000000 1.000000e+00 1.000000e+00
+10: 5.756463 0.000000 3.162278e+02 1.000000e+00
+11: 0.000000 -5.756463 1.000000e+00 3.162278e-03
+12: 0.000000 -11.512925 1.000000e+00 1.000000e-05
+13: -5.756463 -5.756463 3.162278e-03 3.162278e-03
+14: 11.512925 -11.512925 1.000000e+05 1.000000e-05
+15: 5.756463 11.512925 3.162278e+02 1.000000e+05
+16: 0.000000 5.756463 1.000000e+00 3.162278e+02
+17: -5.756463 0.000000 3.162278e-03 1.000000e+00
+18: 5.756463 -11.512925 3.162278e+02 1.000000e-05
+19: -5.756463 11.512925 3.162278e-03 1.000000e+05
+20: 11.512925 11.512925 1.000000e+05 1.000000e+05
+21: 5.756463 -5.756463 3.162278e+02 3.162278e-03
+22: -11.512925 -11.512925 1.000000e-05 1.000000e-05
+23: -11.512925 -5.756463 1.000000e-05 3.162278e-03
+24: 5.756463 5.756463 3.162278e+02 3.162278e+02
+25: -5.756463 5.756463 3.162278e-03 3.162278e+02
+ cost gamma x_domain_cost x_domain_gammaWe plot the performances depending on the evaluated cost and gamma values.
library(ggplot2)
+library(scales)
+autoplot(instance, type = "points", cols_x = c("x_domain_cost", "x_domain_gamma")) +
+ scale_x_continuous(
+ trans = log2_trans(),
+ breaks = trans_breaks("log10", function(x) 10^x),
+ labels = trans_format("log10", math_format(10^.x))) +
+ scale_y_continuous(
+ trans = log2_trans(),
+ breaks = trans_breaks("log10", function(x) 10^x),
+ labels = trans_format("log10", math_format(10^.x)))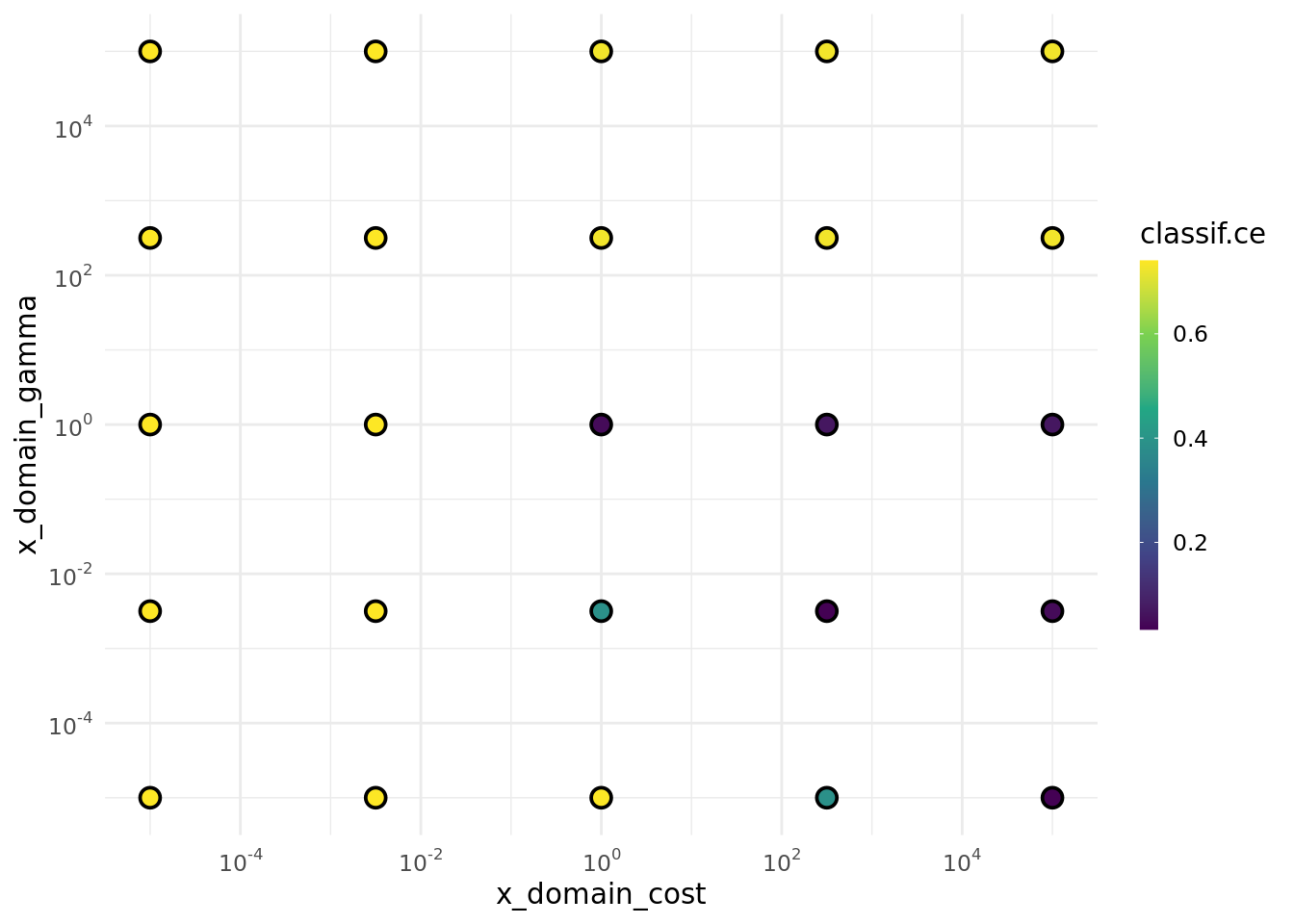
Dependencies ensure that certain parameters are only proposed depending on values of other hyperparameters. We want to tune the degree hyperparameter that is only needed for the polynomial kernel.
learner = lrn("classif.svm", type = "C-classification")
+
+learner$param_set$values$cost = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$gamma = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+
+learner$param_set$values$kernel = to_tune(c("polynomial", "radial"))
+learner$param_set$values$degree = to_tune(1, 4)The dependencies are already stored in the learner parameter set.
+learner$param_set$deps id on cond
+1: cost type <CondEqual[9]>
+2: nu type <CondEqual[9]>
+3: degree kernel <CondEqual[9]>
+4: coef0 kernel <CondAnyOf[9]>
+5: gamma kernel <CondAnyOf[9]>The gamma hyperparameter depends on the kernel being polynomial, radial or sigmoid
learner$param_set$deps$cond[[5]]CondAnyOf: x ∈ {polynomial, radial, sigmoid}whereas the degree hyperparameter is solely used by the polynomial kernel.
learner$param_set$deps$cond[[3]]CondEqual: x = polynomialWe preview the grid to show the effect of the dependencies.
+generate_design_grid(learner$param_set$search_space(), resolution = 2)<Design> with 12 rows:
+ cost gamma kernel degree
+ 1: -11.51293 -11.51293 polynomial 1
+ 2: -11.51293 -11.51293 polynomial 4
+ 3: -11.51293 -11.51293 radial NA
+ 4: -11.51293 11.51293 polynomial 1
+ 5: -11.51293 11.51293 polynomial 4
+ 6: -11.51293 11.51293 radial NA
+ 7: 11.51293 -11.51293 polynomial 1
+ 8: 11.51293 -11.51293 polynomial 4
+ 9: 11.51293 -11.51293 radial NA
+10: 11.51293 11.51293 polynomial 1
+11: 11.51293 11.51293 polynomial 4
+12: 11.51293 11.51293 radial NAThe value for degree is NA if the dependency on the kernel is not satisfied.
We use the tune()-shortcut to run the tuning.
instance = tune(
+ tuner = tnr("grid_search", resolution = 3),
+ task = task,
+ learner = learner,
+ resampling = resampling,
+ measure = measure
+)instance$result cost gamma kernel degree learner_param_vals x_domain classif.ce
+1: 0 11.51293 polynomial 1 <list[5]> <list[4]> 0.02666667We add the optimized hyperparameters to the learner and train the learner on the full dataset.
+learner = lrn("classif.svm")
+learner$param_set$values = instance$result_learner_param_vals
+learner$train(task)The trained model can now be used to make predictions on new data. A common mistake is to report the performance estimated on the resampling sets on which the tuning was performed (instance$result_y) as the model’s performance. These scores might be biased and overestimate the ability of the fitted model to predict with new data. Instead, we have to use nested resampling to get an unbiased performance estimate.
Tuning should not be performed on the same resampling sets which are used for evaluating the model itself, since this would result in a biased performance estimate. Nested resampling uses an outer and inner resampling to separate the tuning from the performance estimation of the model. We can use the AutoTuner class for running nested resampling. The AutoTuner wraps a Learner and tunes the hyperparameter of the learner during $train(). This is our inner resampling loop.
learner = lrn("classif.svm", type = "C-classification")
+learner$param_set$values$cost = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$gamma = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$kernel = to_tune(c("polynomial", "radial"))
+learner$param_set$values$degree = to_tune(1, 4)
+
+resampling_inner = rsmp("cv", folds = 3)
+terminator = trm("none")
+tuner = tnr("grid_search", resolution = 3)
+
+at = AutoTuner$new(
+ learner = learner,
+ resampling = resampling_inner,
+ measure = measure,
+ terminator = terminator,
+ tuner = tuner,
+ store_models = TRUE)We put the AutoTuner into a resample() call to get the outer resampling loop.
resampling_outer = rsmp("cv", folds = 3)
+rr = resample(task = task, learner = at, resampling = resampling_outer, store_models = TRUE)We check the inner tuning results for stable hyperparameters. This means that the selected hyperparameters should not vary too much. We might observe unstable models in this example because the small data set and the low number of resampling iterations might introduce too much randomness. Usually, we aim for the selection of stable hyperparameters for all outer training sets.
+extract_inner_tuning_results(rr)[, .SD, .SDcols = !c("learner_param_vals", "x_domain")] iteration cost gamma kernel degree classif.ce task_id learner_id resampling_id
+1: 1 0.00000 0.00000 polynomial 1 0.03000594 iris classif.svm.tuned cv
+2: 2 0.00000 0.00000 polynomial 1 0.03000594 iris classif.svm.tuned cv
+3: 3 11.51293 -11.51293 radial NA 0.02020202 iris classif.svm.tuned cvNext, we want to compare the predictive performances estimated on the outer resampling to the inner resampling (extract_inner_tuning_results(rr)). Significantly lower predictive performances on the outer resampling indicate that the models with the optimized hyperparameters overfit the data.
rr$score()[, .(iteration, task_id, learner_id, resampling_id, classif.ce)] iteration task_id learner_id resampling_id classif.ce
+1: 1 iris classif.svm.tuned cv 0.04
+2: 2 iris classif.svm.tuned cv 0.06
+3: 3 iris classif.svm.tuned cv 0.00The archives of the AutoTuners allows us to inspect all evaluated hyperparameters configurations with the associated predictive performances.
extract_inner_tuning_archives(rr, unnest = NULL, exclude_columns = c("resample_result", "uhash", "x_domain", "timestamp")) iteration cost gamma kernel degree classif.ce runtime_learners batch_nr warnings errors task_id
+ 1: 1 0.00000 -11.51293 polynomial 2 0.68033274 0.017 1 0 0 iris
+ 2: 1 0.00000 -11.51293 radial NA 0.64111705 0.017 2 0 0 iris
+ 3: 1 0.00000 0.00000 polynomial 1 0.03000594 0.017 3 0 0 iris
+ 4: 1 11.51293 0.00000 polynomial 1 0.05050505 0.012 4 0 0 iris
+ 5: 1 11.51293 11.51293 radial NA 0.66013072 0.014 5 0 0 iris
+ ---
+104: 3 11.51293 -11.51293 polynomial 2 0.68924540 0.013 32 0 0 iris
+105: 3 -11.51293 -11.51293 radial NA 0.68924540 0.012 33 0 0 iris
+106: 3 -11.51293 11.51293 polynomial 4 0.11883541 0.015 34 0 0 iris
+107: 3 0.00000 -11.51293 polynomial 4 0.68924540 0.015 35 0 0 iris
+108: 3 11.51293 11.51293 polynomial 1 0.06951872 0.725 36 0 0 iris
+ learner_id resampling_id
+ 1: classif.svm.tuned cv
+ 2: classif.svm.tuned cv
+ 3: classif.svm.tuned cv
+ 4: classif.svm.tuned cv
+ 5: classif.svm.tuned cv
+ ---
+104: classif.svm.tuned cv
+105: classif.svm.tuned cv
+106: classif.svm.tuned cv
+107: classif.svm.tuned cv
+108: classif.svm.tuned cvThe aggregated performance of all outer resampling iterations is essentially the unbiased performance of an SVM with optimal hyperparameter found by grid search.
+rr$aggregate()classif.ce
+0.03333333 Applying nested resampling can be shortened by using the tune_nested()-shortcut.
learner = lrn("classif.svm", type = "C-classification")
+learner$param_set$values$cost = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$gamma = to_tune(p_dbl(1e-5, 1e5, logscale = TRUE))
+learner$param_set$values$kernel = to_tune(c("polynomial", "radial"))
+learner$param_set$values$degree = to_tune(1, 4)
+
+rr = tune_nested(
+ tuner = tnr("grid_search", resolution = 3),
+ task = tsk("iris"),
+ learner = learner,
+ inner_resampling = rsmp ("cv", folds = 3),
+ outer_resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.ce"),
+)The mlr3book includes chapters on tuning spaces and hyperparameter tuning. The mlr3cheatsheets contain frequently used commands and workflows of mlr3.
+ + + +# include: false
+requireNamespace("bst")Loading required namespace: bstrequireNamespace("fastICA")Loading required namespace: fastICAIn this use case we show how to tune a rather complex graph consisting of different preprocessing steps and different learners where each preprocessing step and learner itself has parameters that can be tuned. You will learn the following:
+Graph that consists of two common preprocessing steps, then switches between two dimensionality reduction techniques followed by a Learner vs. no dimensionality reduction followed by another Learnergrid search to find an optimal choice of preprocessing steps and hyperparameters.Ideally you already had a look at how to tune over multiple learners.
+First, we load the packages we will need:
+library(mlr3verse)
+library(mlr3learners)We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented. The lgr package is used for logging in all mlr3 packages. The mlr3 logger prints the logging messages from the base package, whereas the bbotk logger is responsible for logging messages from the optimization packages (e.g. mlr3tuning ).
set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")We are going to work with some gene expression data included as a supplement in the bst package. The data consists of 2308 gene profiles in 63 training and 20 test samples. The following data preprocessing steps are done analogously as in vignette("khan", package = "bst"):
datafile = system.file("extdata", "supplemental_data", package = "bst")
+dat0 = read.delim(datafile, header = TRUE, skip = 1)[, -(1:2)]
+dat0 = t(dat0)
+dat = data.frame(dat0[!(rownames(dat0) %in%
+ c("TEST.9", "TEST.13", "TEST.5", "TEST.3", "TEST.11")), ])
+dat$class = as.factor(
+ c(substr(rownames(dat)[1:63], start = 1, stop = 2),
+ c("NB", "RM", "NB", "EW", "RM", "BL", "EW", "RM", "EW", "EW", "EW", "RM",
+ "BL", "RM", "NB", "NB", "NB", "NB", "BL", "EW")
+ )
+)We then construct our training and test Task :
task = as_task_classif(dat, target = "class", id = "SRBCT")
+task_train = task$clone(deep = TRUE)
+task_train$filter(1:63)
+task_test = task$clone(deep = TRUE)
+task_test$filter(64:83)Our graph will start with log transforming the features, followed by scaling them. Then, either a PCA or ICA is applied to extract principal / independent components followed by fitting a LDA or a ranger random forest is fitted without any preprocessing (the log transformation and scaling should most likely affect the LDA more than the ranger random forest). Regarding the PCA and ICA, both the number of principal / independent components are tuning parameters. Regarding the LDA, we can further choose different methods for estimating the mean and variance and regarding the ranger, we want to tune the mtry and num.tree parameters. Note that the PCA-LDA combination has already been successfully applied in different cancer diagnostic contexts when the feature space is of high dimensionality (Morais and Lima 2018).
To allow for switching between the PCA / ICA-LDA and ranger we can either use branching or proxy pipelines, i.e., PipeOpBranch and PipeOpUnbranch or PipeOpProxy. We will first cover branching in detail and later show how the same can be done using PipeOpProxy.
First, we have a look at the baseline classification accuracy of the LDA and ranger on the training task:
base = benchmark(benchmark_grid(
+ task_train,
+ learners = list(lrn("classif.lda"), lrn("classif.ranger")),
+ resamplings = rsmp("cv", folds = 3)))Warning in lda.default(x, grouping, ...): variables are collinear
+
+Warning in lda.default(x, grouping, ...): variables are collinear
+
+Warning in lda.default(x, grouping, ...): variables are collinearbase$aggregate(measures = msr("classif.acc")) nr task_id learner_id resampling_id iters classif.acc
+1: 1 SRBCT classif.lda cv 3 0.6666667
+2: 2 SRBCT classif.ranger cv 3 0.9206349
+Hidden columns: resample_resultThe out-of-the-box ranger appears to already have good performance on the training task. Regarding the LDA, we do get a warning message that some features are colinear. This strongly suggests to reduce the dimensionality of the feature space. Let’s see if we can get some better performance, at least for the LDA.
Our graph starts with log transforming the features (we explicitly use base 10 only for better interpretability when inspecting the model later), using PipeOpColApply, followed by scaling the features using PipeOpScale. Then, the first branch allows for switching between the PCA / ICA-LDA and ranger, and within PCA / ICA-LDA, the second branch allows for switching between PCA and ICA:
graph1 =
+ po("colapply", applicator = function(x) log(x, base = 10)) %>>%
+ po("scale") %>>%
+ # pca / ica followed by lda vs. ranger
+ po("branch", id = "branch_learner", options = c("pca_ica_lda", "ranger")) %>>%
+ gunion(list(
+ po("branch", id = "branch_preproc_lda", options = c("pca", "ica")) %>>%
+ gunion(list(
+ po("pca"), po("ica")
+ )) %>>%
+ po("unbranch", id = "unbranch_preproc_lda") %>>%
+ lrn("classif.lda"),
+ lrn("classif.ranger")
+ )) %>>%
+ po("unbranch", id = "unbranch_learner")Note that the names of the options within each branch are arbitrary, but ideally they describe what is happening. Therefore we go with "pca_ica_lda" / "ranger” and "pca" / "ica". Finally, we also could have used the branch ppl to make branching easier (we will come back to this in the Proxy section). The graph looks like the following:
graph1$plot(html = FALSE)
We can inspect the parameters of the ParamSet of the graph to see which parameters can be set:
graph1$param_set$ids() [1] "colapply.applicator" "colapply.affect_columns"
+ [3] "scale.center" "scale.scale"
+ [5] "scale.robust" "scale.affect_columns"
+ [7] "branch_learner.selection" "branch_preproc_lda.selection"
+ [9] "pca.center" "pca.scale."
+[11] "pca.rank." "pca.affect_columns"
+[13] "ica.n.comp" "ica.alg.typ"
+[15] "ica.fun" "ica.alpha"
+[17] "ica.method" "ica.row.norm"
+[19] "ica.maxit" "ica.tol"
+[21] "ica.verbose" "ica.w.init"
+[23] "ica.affect_columns" "classif.lda.dimen"
+[25] "classif.lda.method" "classif.lda.nu"
+[27] "classif.lda.predict.method" "classif.lda.predict.prior"
+[29] "classif.lda.prior" "classif.lda.tol"
+[31] "classif.ranger.alpha" "classif.ranger.always.split.variables"
+[33] "classif.ranger.class.weights" "classif.ranger.holdout"
+[35] "classif.ranger.importance" "classif.ranger.keep.inbag"
+[37] "classif.ranger.max.depth" "classif.ranger.min.node.size"
+[39] "classif.ranger.min.prop" "classif.ranger.minprop"
+[41] "classif.ranger.mtry" "classif.ranger.mtry.ratio"
+[43] "classif.ranger.num.random.splits" "classif.ranger.num.threads"
+[45] "classif.ranger.num.trees" "classif.ranger.oob.error"
+[47] "classif.ranger.regularization.factor" "classif.ranger.regularization.usedepth"
+[49] "classif.ranger.replace" "classif.ranger.respect.unordered.factors"
+[51] "classif.ranger.sample.fraction" "classif.ranger.save.memory"
+[53] "classif.ranger.scale.permutation.importance" "classif.ranger.se.method"
+[55] "classif.ranger.seed" "classif.ranger.split.select.weights"
+[57] "classif.ranger.splitrule" "classif.ranger.verbose"
+[59] "classif.ranger.write.forest" The id’s are prefixed by the respective PipeOp they belong to, e.g., pca.rank. refers to the rank. parameter of PipeOpPCA.
Our graph either fits a LDA after applying PCA or ICA, or alternatively a ranger with no preprocessing. These two options each define selection parameters that we can tune. Moreover, within the respective PipeOp’s we want to tune the following parameters: pca.rank., ica.n.comp, classif.lda.method, classif.ranger.mtry, and classif.ranger.num.trees. The first two parameters are integers that in-principal could range from 1 to the number of features. However, for ICA, the upper bound must not exceed the number of observations and as we will later use 3-fold cross-validation as the resampling method for the tuning, we just set the upper bound to 30 (and do the same for PCA). Regarding the classif.lda.method we will only be interested in "moment" estimation vs. minimum volume ellipsoid covariance estimation ("mve"). Moreover, we set the lower bound of classif.ranger.mtry to 200 (which is around the number of features divided by 10) and the upper bound to 1000.
tune_ps1 = ps(
+ branch_learner.selection =
+ p_fct(c("pca_ica_lda", "ranger")),
+ branch_preproc_lda.selection =
+ p_fct(c("pca", "ica"), depends = branch_learner.selection == "pca_ica_lda"),
+ pca.rank. =
+ p_int(1, 30, depends = branch_preproc_lda.selection == "pca"),
+ ica.n.comp =
+ p_int(1, 30, depends = branch_preproc_lda.selection == "ica"),
+ classif.lda.method =
+ p_fct(c("moment", "mve"), depends = branch_preproc_lda.selection == "ica"),
+ classif.ranger.mtry =
+ p_int(200, 1000, depends = branch_learner.selection == "ranger"),
+ classif.ranger.num.trees =
+ p_int(500, 2000, depends = branch_learner.selection == "ranger"))The parameter branch_learner.selection defines whether we go down the left (PCA / ICA followed by LDA) or the right branch (ranger). The parameter branch_preproc_lda.selection defines whether a PCA or ICA will be applied prior to the LDA. The other parameters directly belong to the ParamSet of the PCA / ICA / LDA / ranger. Note that it only makes sense to switch between PCA / ICA if the "pca_ica_lda" branch was selected beforehand. We have to specify this via the depends parameter.
Finally, we also could have proceeded to tune the numeric parameters on a log scale. I.e., looking at pca.rank. the performance difference between rank 1 and 2 is probably much larger than between rank 29 and rank 30. The mlr3tuning Tutorial covers such transformations.
We can now tune the parameters of our graph as defined in the search space with respect to a measure. We will use the classification accuracy. As a resampling method we use 3-fold cross-validation. We will use the TerminatorNone (i.e., no early termination) for terminating the tuning because we will apply a grid search (we use a grid search because it gives nicely plottable and understandable results but if there were much more parameters, random search or more intelligent optimization methods would be preferred to a grid search:
tune1 = TuningInstanceSingleCrit$new(
+ task_train,
+ learner = graph1,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.acc"),
+ search_space = tune_ps1,
+ terminator = trm("none")
+)We then perform a grid search using a resolution of 4 for the numeric parameters. The grid being used will look like the following (note that the dependencies we specified above are handled automatically):
generate_design_grid(tune_ps1, resolution = 4)We trigger the tuning.
+tuner_gs = tnr("grid_search", resolution = 4, batch_size = 10)
+tuner_gs$optimize(tune1) branch_learner.selection branch_preproc_lda.selection pca.rank. ica.n.comp classif.lda.method classif.ranger.mtry
+1: pca_ica_lda ica NA 10 mve NA
+ classif.ranger.num.trees learner_param_vals x_domain classif.acc
+1: NA <list[8]> <list[4]> 0.984127Now, we can inspect the results ordered by the classification accuracy:
as.data.table(tune1$archive)[order(classif.acc), ]We achieve very good accuracy using ranger, more or less regardless how mtry and num.trees are set. However, the LDA also shows very good accuracy when combined with PCA or ICA retaining 30 components.
For now, we decide to use ranger with mtry set to 200 and num.trees set to 1000.
Setting these parameters manually in our graph, then training on the training task and predicting on the test task yields an accuracy of:
+graph1$param_set$values$branch_learner.selection = "ranger"
+graph1$param_set$values$classif.ranger.mtry = 200
+graph1$param_set$values$classif.ranger.num.trees = 1000
+graph1$train(task_train)$unbranch_learner.output
+NULLgraph1$predict(task_test)[[1L]]$score(msr("classif.acc"))classif.acc
+ 1 Note that we also could have wrapped our graph in a GraphLearner and proceeded to use this as a learner in an AutoTuner.
Instead of using branches to split our graph with respect to the learner and preprocessing options, we can also use PipeOpProxy. PipeOpProxy accepts a single content parameter that can contain any other PipeOp or Graph. This is extremely flexible in the sense that we do not have to specify our options during construction. However, the parameters of the contained PipeOp or Graph are no longer directly contained in the ParamSet of the resulting graph. Therefore, when tuning the graph, we do have to make use of a trafo function.
graph2 =
+ po("colapply", applicator = function(x) log(x, base = 10)) %>>%
+ po("scale") %>>%
+ po("proxy")This graph now looks like the following:
+graph2$plot(html = FALSE)
At first, this may look like a linear graph. However, as the content parameter of PipeOpProxy can be tuned and set to contain any other PipeOp or Graph, this will allow for a similar non-linear graph as when doing branching.
graph2$param_set$ids()[1] "colapply.applicator" "colapply.affect_columns" "scale.center" "scale.scale"
+[5] "scale.robust" "scale.affect_columns" "proxy.content" We can tune the graph by using the same search space as before. However, here the trafo function is of central importance to actually set our options and parameters:
tune_ps2 = tune_ps1$clone(deep = TRUE)The trafo function does all the work, i.e., selecting either the PCA / ICA-LDA or ranger as the proxy.content as well as setting the parameters of the respective preprocessing PipeOps and Learners.
proxy_options = list(
+ pca_ica_lda =
+ ppl("branch", graphs = list(pca = po("pca"), ica = po("ica"))) %>>%
+ lrn("classif.lda"),
+ ranger = lrn("classif.ranger")
+)Above, we made use of the branch ppl allowing us to easily construct a branching graph. Of course we also could have use another nested PipeOpProxy to specify the preprocessing options ("pca" vs. "ica") within proxy_options if for some reason we do not want to do branching at all. The trafo function below selects one of the proxy_options from above and sets the respective parameters for the PCA, ICA, LDA and ranger. Here, the argument x is a list which will contain sampled / selected parameters from our ParamSet (in our case, tune_ps2). The return value is a list only including the appropriate proxy.content parameter. In each tuning iteration, the proxy.content parameter of our graph will be set to this value.
tune_ps2$trafo = function(x, param_set) {
+ proxy.content = proxy_options[[x$branch_learner.selection]]
+ if (x$branch_learner.selection == "pca_ica_lda") {
+ # pca_ica_lda
+ proxy.content$param_set$values$branch.selection = x$branch_preproc_lda.selection
+ if (x$branch_preproc_lda.selection == "pca") {
+ proxy.content$param_set$values$pca.rank. = x$pca.rank.
+ } else {
+ proxy.content$param_set$values$ica.n.comp = x$ica.n.comp
+ }
+ proxy.content$param_set$values$classif.lda.method = x$classif.lda.method
+ } else {
+ # ranger
+ proxy.content$param_set$values$mtry = x$classif.ranger.mtry
+ proxy.content$param_set$values$num.trees = x$classif.ranger.num.trees
+ }
+ list(proxy.content = proxy.content)
+}I.e., suppose that the following parameters will be selected from our ParamSet:
x = list(
+ branch_learner.selection = "ranger",
+ classif.ranger.mtry = 200,
+ classif.ranger.num.trees = 500)The trafo function will then return:
tune_ps2$trafo(x)$proxy.content
+<LearnerClassifRanger:classif.ranger>
+* Model: -
+* Parameters: num.threads=1, mtry=200, num.trees=500
+* Packages: mlr3, mlr3learners, ranger
+* Predict Types: [response], prob
+* Feature Types: logical, integer, numeric, character, factor, ordered
+* Properties: hotstart_backward, importance, multiclass, oob_error, twoclass, weightsTuning can be carried out analogously as done above:
+tune2 = TuningInstanceSingleCrit$new(
+ task_train,
+ learner = graph2,
+ resampling = rsmp("cv", folds = 3),
+ measure = msr("classif.acc"),
+ search_space = tune_ps2,
+ terminator = trm("none")
+)
+tuner_gs$optimize(tune2)as.data.table(tune2$archive)[order(classif.acc), ]Tuner for real-valued search spaces are not able to tune on integer hyperparameters. However, it is possible to round the real values proposed by a Tuner to integers before passing them to the learner in the evaluation. We show how to apply a parameter transformation to a ParamSet and use this set in the tuning process.
We load the mlr3verse package which pulls in the most important packages for this example.
+library(mlr3verse)Loading required package: mlr3We initialize the random number generator with a fixed seed for reproducibility, and decrease the verbosity of the logger to keep the output clearly represented.
+set.seed(7832)
+lgr::get_logger("mlr3")$set_threshold("warn")
+lgr::get_logger("bbotk")$set_threshold("warn")In this example, we use the k-Nearest-Neighbor classification learner. We want to tune the integer-valued hyperparameter k which defines the numbers of neighbors.
learner = lrn("classif.kknn")
+print(learner$param_set$params$k) id class lower upper levels default
+1: k ParamInt 1 Inf 7We choose generalized simulated annealing as tuning strategy. The param_classes field of TunerGenSA states that the tuner only supports real-valued (ParamDbl) hyperparameter tuning.
print(tnr("gensa"))<TunerGenSA>: Generalized Simulated Annealing
+* Parameters: trace.mat=FALSE, smooth=FALSE
+* Parameter classes: ParamDbl
+* Properties: single-crit
+* Packages: mlr3tuning, bbotk, GenSATo get integer-valued hyperparameter values for k, we construct a search space with a transformation function. The as.integer() function converts any real valued number to an integer by removing the decimal places.
search_space = ps(
+ k = p_dbl(lower = 3, upper = 7.99, trafo = as.integer)
+)We start the tuning and compare the results of the search space to the results in the space of the learners hyperparameter set.
+instance = tune(
+ tuner = tnr("gensa"),
+ task = tsk("iris"),
+ learner = learner,
+ resampling = rsmp("holdout"),
+ measure = msr("classif.ce"),
+ term_evals = 20,
+ search_space = search_space)Warning in optim(theta.old, fun, gradient, control = control, method = method, : one-dimensional optimization by Nelder-Mead is unreliable:
+use "Brent" or optimize() directlyThe optimal k is still a real number in the search space.
instance$result_x_search_space k
+1: 3.82686However, in the learners hyperparameters space, k is an integer value.
instance$result_x_domain$k
+[1] 3The archive shows us that for all real-valued k proposed by GenSA, an integer-valued k in the learner hyperparameter space (x_domain_k) was created.
as.data.table(instance$archive)[, .(k, classif.ce, x_domain_k)] k classif.ce x_domain_k
+ 1: 3.826860 0.06 3
+ 2: 5.996323 0.06 5
+ 3: 5.941332 0.06 5
+ 4: 3.826860 0.06 3
+ 5: 3.826860 0.06 3
+ 6: 3.826860 0.06 3
+ 7: 4.209546 0.06 4
+ 8: 3.444174 0.06 3
+ 9: 4.018203 0.06 4
+10: 3.635517 0.06 3
+11: 3.922532 0.06 3
+12: 3.731189 0.06 3
+13: 3.874696 0.06 3
+14: 3.779024 0.06 3
+15: 3.850778 0.06 3
+16: 3.802942 0.06 3
+17: 3.838819 0.06 3
+18: 3.814901 0.06 3
+19: 3.832840 0.06 3
+20: 3.820881 0.06 3Internally, TunerGenSA was given the parameter types of the search space and therefore suggested real numbers for k. Before the performance of the different k values was evaluated, the transformation function of the search_space parameter set was called and k was transformed to an integer value.
Note that the tuner is not aware of the transformation. This has two problematic consequences: First, the tuner might propose different real valued configurations that after rounding end up to be already evaluated configurations and we end up with re-evaluating the same hyperparameter configuration. This is only problematic, if we only optimze integer parameters. Second, the rounding introduces discontinuities which can be problematic for some tuners.
+We successfully tuned a integer-valued hyperparameter with TunerGenSA which is only suitable for an real-valued search space. This technique is not limited to tuning problems. Optimizer in bbotk can be also used in the same way to produce points with integer parameters.
${w.content.map(p).join("")}
`:""}return f.map(p).join("")}function LRt(i,a){a&&i.attr("style",a)}function MRt(i,a,f,p){const w=i.append("foreignObject"),y=w.append("xhtml:div"),b=a.label,E=a.isNode?"nodeLabel":"edgeLabel";y.html(`"+b+""),LRt(y,a.labelStyle),y.style("display","table-cell"),y.style("white-space","nowrap"),y.style("max-width",f+"px"),y.attr("xmlns","http://www.w3.org/1999/xhtml");let S=y.node().getBoundingClientRect();return S.width===f&&(y.style("display","table"),y.style("white-space","break-spaces"),y.style("width",f+"px"),S=y.node().getBoundingClientRect()),w.style("width",S.width),w.style("height",S.height),w.node()}function IBe(i,a,f){return i.append("tspan").attr("class","text-outer-tspan").attr("x",0).attr("y",a*f-.1+"em").attr("dy",f+"em")}function DRt(i,a,f,p=!1){const y=a.append("g");let b=y.insert("rect").attr("class","background");const E=y.append("text").attr("y","-10.1");let S=-1;if(f.forEach(N=>{S++;let B=IBe(E,S,1.1),R=[...N].reverse(),j,$=[];for(;R.length;)j=R.pop(),$.push(j),OBe(B,$),B.node().getComputedTextLength()>i&&($.pop(),R.push(j),OBe(B,$),$=[],S++,B=IBe(E,S,1.1))}),p){const N=E.node().getBBox(),B=2;return b.attr("x",-B).attr("y",-B).attr("width",N.width+2*B).attr("height",N.height+2*B),y.node()}else return E.node()}function OBe(i,a){i.text(""),a.forEach((f,p)=>{const w=i.append("tspan").attr("font-style",f.type==="em"?"italic":"normal").attr("class","text-inner-tspan").attr("font-weight",f.type==="strong"?"bold":"normal");p===0?w.text(f.content):w.text(" "+f.content)})}const dK=(i,a="",{style:f="",isTitle:p=!1,classes:w="",useHtmlLabels:y=!0,isNode:b=!0,width:E,addSvgBackground:S=!1}={})=>{if(Fe.info("createText",a,f,p,w,y,b,S),y){const N=ARt(a),B={isNode:b,label:XN(N).replace(/fa[blrs]?:fa-[\w-]+/g,j=>``),labelStyle:f.replace("fill:","color:")};return MRt(i,B,E,w)}else{const N=SRt(a),B=['"',"'",".",",",":",";","!","?","(",")","[","]","{","}"];let R;return N.forEach($=>{$.forEach(V=>{B.includes(V.content)&&R&&(R.content+=V.content,V.content=""),R=V})}),DRt(E,i,N,S)}},jd=async(i,a,f,p)=>{let w;const y=a.useHtmlLabels||d1(Tt().flowchart.htmlLabels);f?w=f:w="node default";const b=i.insert("g").attr("class",w).attr("id",a.domId||a.id),E=b.insert("g").attr("class","label").attr("style",a.labelStyle);let S;a.labelText===void 0?S="":S=typeof a.labelText=="string"?a.labelText:a.labelText[0];const N=E.node();let B;a.labelType==="markdown"?B=dK(E,ep(XN(S),Tt()),{useHtmlLabels:y,width:a.width||Tt().flowchart.wrappingWidth,classes:"markdown-node-label"}):B=N.appendChild(sp(ep(XN(S),Tt()),a.labelStyle,!1,p));let R=B.getBBox();const j=a.padding/2;if(d1(Tt().flowchart.htmlLabels)){const $=B.children[0],V=Cr(B),Q=$.getElementsByTagName("img");if(Q){const oe=S.replace(/