-
Notifications
You must be signed in to change notification settings - Fork 6
/
Copy pathREADME.Rmd
941 lines (788 loc) · 45.2 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
816
817
818
819
820
821
822
823
824
825
826
827
828
829
830
831
832
833
834
835
836
837
838
839
840
841
842
843
844
845
846
847
848
849
850
851
852
853
854
855
856
857
858
859
860
861
862
863
864
865
866
867
868
869
870
871
872
873
874
875
876
877
878
879
880
881
882
883
884
885
886
887
888
889
890
891
892
893
894
895
896
897
898
899
900
901
902
903
904
905
906
907
908
909
910
911
912
913
914
915
916
917
918
919
920
921
922
923
924
925
926
927
928
929
930
931
932
933
934
935
936
937
938
939
940
941
---
output:
github_document:
toc: false
toc_depth: 1
editor_options:
chunk_output_type: console
---
<!-- README.md is generated from README.Rmd -->
```{r setup, include=FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
eval = FALSE,
comment = "#>",
fig.path = "README-",
out.width = "100%"
)
```
<!-- badges: start -->
[](https://cran.r-project.org/package=ctrdata)
[](https://app.codecov.io/gh/rfhb/ctrdata)
[](https://github.com/rfhb/ctrdata/actions/workflows/check-standard-win-macos-linux.yaml)
<!-- badges: end -->
[Main features](#main-features) •
[References](#references) •
[Installation](#installation) •
[Overview](#overview-of-functions-in-ctrdata) •
[Databases](#databases-for-use-with-ctrdata) •
[Data model](#data-model-of-ctrdata) •
[Example workflow](#example-workflow) •
[Analysis across trials](#workflow-cross-trial-example) •
[Tests](#tests) •
[Acknowledgements](#acknowledgements) •
[Future](#future-features)
# ctrdata for aggregating and analysing clinical trials
The package `ctrdata` provides functions for retrieving (downloading), aggregating and analysing clinical trials using information (structured protocol and result data, as well as documents) from public registers. It can be used with the
- EU Clinical Trials Register ("EUCTR", https://www.clinicaltrialsregister.eu/)
- EU Clinical Trials Information System ("CTIS", https://euclinicaltrials.eu/, see [example](#workflow-ctis-example))
- ClinicalTrials.gov ("CTGOV2", see [example](#workflow-ctgov-example))
- ISRCTN Registry (https://www.isrctn.com/)
The motivation is to investigate the design and conduct of trials of interest, to describe their trends and availability for patients and to facilitate using their detailed results for research and meta-analyses. `ctrdata` is a package for the [R](https://www.r-project.org/) system, but other systems and tools can use the databases created with this package. This README was reviewed on 2024-12-27 for version 1.20.0.9000.
## Main features
* Protocol- and results-related trial information is easily downloaded: Users define a query in a register's web interface, then copy the URL and enter it into `ctrdata` which retrieves in one go all trials found. A [script](#2-script-to-automatically-copy-users-query-from-web-browser) can automate copying the query URL from all registers. Personal annotations can be made when downloading trials. Also, [trial documents](#documents-example) and [historic versions](https://rfhb.github.io/ctrdata/articles/ctrdata_summarise.html#analysing-sample-size-using-historic-versions-of-trial-records) as available in registers on trials can be downloaded.
* Downloaded trial information is transformed and stored in a collection of a document-centric database, for fast and offline access. Information from different registers can be accumulated in a single collection. Uses `DuckDB`, `PostgreSQL`, `RSQLite` or `MongoDB`, via R package `nodbi`: see section [Databases](#databases-that-can-be-used-with-ctrdata) below. Interactively browse through trial structure and data. Easily re-run any previous query in a collection to retrieve and update trial records.
* For analyses, convenience functions in `ctrdata` allow find synonyms of an active substance, to identify unique (de-duplicated) trial records across all registers, to merge and recode fields as well as to easily access deeply-nested fields. Analysis can be done with `R` (see [vignette](https://rfhb.github.io/ctrdata/articles/ctrdata_summarise.html)) or other systems, using the `JSON`-[structured information in the database](#trial-records-in-databases).
Remember to respect the registers' terms and conditions (see `ctrOpenSearchPagesInBrowser(copyright = TRUE)`). Please cite this package in any publication as follows: "Ralf Herold (2024). _ctrdata: Retrieve and Analyze Clinical Trials in Public Registers._ R package version 1.20.0, https://cran.r-project.org/package=ctrdata".
<!--
```{r}
citation("ctrdata")
# find publications
utils::browseURL("https://scholar.google.com/scholar?hl=en&as_sdt=0,5&q=%22ctrdata%22")
# mentioned but not used in
# * Sciannameo et al. (2024) Information Extraction from Medical Case Reports Using OpenAI InstructGPT. Computer Methods and Programs in Biomedicine [https://doi.org/10.1016/j.cmpb.2024.108326](https://doi.org/10.1016/j.cmpb.2024.108326)
```
-->
## References
Package `ctrdata` has been used for unpublished works and for these publications:
* Alzheimer’s disease Horizon Scanning Report (2024) [PDF file, p 10](https://www.ema.europa.eu/en/documents/report/alzheimers-disease-eu-horizon-scanning-report_en.pdf#page=10) `r emoji::emoji("bell")`
* Kundu et al. (2024) Analysis of Factors Influencing Enrollment Success in Hematology Malignancy Cancer Clinical Trials (2008-2023). Blood Meeting Abstracts [https://doi.org/10.1182/blood-2024-207446](https://doi.org/10.1182/blood-2024-207446) `r emoji::emoji("bell")`
* Lasch et al. (2022) The Impact of COVID‐19 on the Initiation of Clinical Trials in Europe and the United States. Clinical Pharmacology & Therapeutics [https://doi.org/10.1002/cpt.2534](https://doi.org/10.1002/cpt.2534)
* Sood et al. (2022) Managing the Evidence Infodemic: Automation Approaches Used for Developing NICE COVID-19 Living Guidelines. medRxiv [https://doi.org/10.1101/2022.06.13.22276242](https://doi.org/10.1101/2022.06.13.22276242)
* Blogging on [Innovation coming to paediatric research](https://paediatricdata.eu/innovation-coming-to-paediatric-research/)
* Cancer Research UK (2017) [The impact of collaboration: The value of UK medical research to EU science and health](https://www.cancerresearchuk.org/about-us/we-develop-policy/policy-publications-and-research-tenders#Policy_publications4)
* EMA (2017) Results of juvenile animal studies (JAS) and impact on anti-cancer medicine development and use in children [PDF file, p 34](https://www.ema.europa.eu/en/documents/scientific-guideline/results-juvenile-animal-studies-jas-and-impact-anti-cancer-medicine-development-and-use-children_en.pdf#page=34)
## Installation
### 1. Install package `ctrdata` in R
Package `ctrdata` is [on CRAN](https://cran.r-project.org/package=ctrdata) and [on GitHub](https://github.com/rfhb/ctrdata). Within [R](https://www.r-project.org/), use the following commands to install package `ctrdata`:
```{r install_ctrdata, eval=FALSE}
# Install CRAN version:
install.packages("ctrdata")
# Alternatively, install development version:
install.packages("devtools")
devtools::install_github("rfhb/ctrdata", build_vignettes = TRUE)
```
These commands also install the package's dependencies (`jsonlite`, `httr`, `curl`, `clipr`, `xml2`, `nodbi`, `stringi`, `tibble`, `lubridate`, `jqr`, `dplyr`, `zip` and `V8`).
### 2. Script to automatically copy user's query from web browser
This is optional; it works with all registers supported by `ctrdata` but is recommended for CTIS because the URL in the web browser does not reflect the parameters the user specified for querying this register.
In the web browser, install the [Tampermonkey browser extension](https://www.tampermonkey.net/), click on "New user script" and then on "Tools", enter into "Import from URL" this URL: [`https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js`](https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js) and then click on "Install".
The browser extension can be disabled and enabled by the user. When enabled, the URLs to all user's queries in the registers are automatically copied to the clipboard and can be pasted into the `queryterm = ...` parameter of function [ctrLoadQueryIntoDb()](https://rfhb.github.io/ctrdata/reference/ctrLoadQueryIntoDb.html).
Additionally, this script retrieves results for `CTIS` search URLs such as [https://euclinicaltrials.eu/ctis-public/search#searchCriteria={"status":[3,4]}](https://euclinicaltrials.eu/ctis-public/search#searchCriteria={"status":[3,4]}).
## Overview of functions in `ctrdata`
The functions are listed in the approximate order of use in a user's workflow (in bold, main functions). See also the [package documentation overview](https://rfhb.github.io/ctrdata/reference/index.html).
Function name | Function purpose
---------------------------- | --------------------------------------------
`ctrOpenSearchPagesInBrowser()` | Open search pages of registers or execute search in web browser
`ctrFindActiveSubstanceSynonyms()` | Find synonyms and alternative names for an active substance
`ctrGetQueryUrl()` | Import from clipboard the URL of a search in one of the registers
`ctrLoadQueryIntoDb()` | **Retrieve (download) or update, and annotate, information on trials from a register and store in a collection in a database**
`ctrShowOneTrial()` | `r emoji::emoji("bell")` Show full structure and all data of a trial, interactively select fields of interest for `dbGetFieldsIntoDf()`
`dbQueryHistory()` | Show the history of queries that were downloaded into the collection
`dbFindIdsUniqueTrials()` | **Get the identifiers of de-duplicated trials in the collection**
`dbFindFields()` | Find names of variables (fields) in the collection
`dbGetFieldsIntoDf()` | **Create a data frame (or tibble) from trial records in the database with the specified fields**
`dfTrials2Long()` | Transform the data.frame from `dbGetFieldsIntoDf()` into a long name-value data.frame, including deeply nested fields
`dfName2Value()` | From a long name-value data.frame, extract values for variables (fields) of interest (e.g., endpoints)
`dfMergeVariablesRelevel()` | Merge variables into a new variable, optionally map values to a new set of levels
## Databases for use with `ctrdata`
Package `ctrdata` retrieves trial information and stores it in a database collection, which has to be given as a connection object to parameter `con` for several `ctrdata` functions; this connection object is created in slightly different ways for the four supported database backends that can be used with `ctrdata` as shown in the table. For a speed comparison, see the [nodbi documentation](https://github.com/ropensci/nodbi#benchmark).
Besides ctrdata functions below, any such a connection object can equally be used with functions of other packages, such as `nodbi` (last row in table) or, in case of MongoDB as database backend, `mongolite` (see vignettes).
Purpose | Function call
-------------------- | --------------------
Create **SQLite** database connection | `dbc <- nodbi::src_sqlite(dbname = "name_of_my_database", collection = "name_of_my_collection")`
Create **MongoDB** database connection | `dbc <- nodbi::src_mongo(db = "name_of_my_database", collection = "name_of_my_collection")`
Create **PostgreSQL** database connection | `dbc <- nodbi::src_postgres(dbname = "name_of_my_database"); dbc[["collection"]] <- "name_of_my_collection"`
Create **DuckDB** database connection | `dbc <- nodbi::src_duckdb(dbdir = "name_of_my_database", collection = "name_of_my_collection")`
Use connection with `ctrdata` functions | `ctrdata::{ctrLoadQueryIntoDb, dbQueryHistory, dbFindIdsUniqueTrials, dbFindFields, dbGetFieldsIntoDf}(con = dbc, ...)`
Use connection with `nodbi` functions | e.g., `nodbi::docdb_query(src = dbc, key = dbc$collection, ...)`
## Data model of `ctrdata`
Package `ctrdata` uses the data models that are implicit in data retrieved from the different registers. No mapping is provided for any register's data model to a putative target data model. The reasons include that registers' data models are notably evolving over time and that there are only few data fields with similar values and meaning between the registers.
Thus, the handling of data from different models of registers is to be done at the time of analysis. This approach allows a high level of flexibility, transparency and reproducibility. See examples in the help text for function [dfMergeVariablesRelevel()](https://rfhb.github.io/ctrdata/reference/dfMergeVariablesRelevel.html) and section [Analysis across trials](#workflow-cross-trial-example) below for how to align related fields from different registers for a joint analysis.
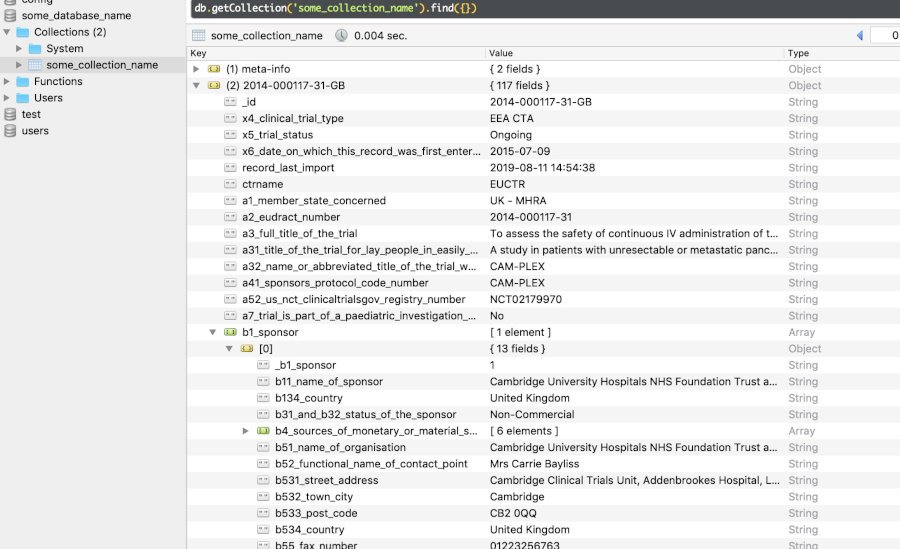
In any of the `NoSQL` [databases](https://rfhb.github.io/ctrdata/index.html#databases-for-use-with-ctrdata), one clinical trial is one document, corresponding to one row in a `SQLite`, `PostgreSQL` or `DuckDB` table, and to one document in a `MongoDB` collection. The `NoSQL` backends allow documents to have different structures, which is used here to accommodate the different data models of registers. Package `ctrdata` stores in every such document:
- field `_id` with the trial identification as provided by the register from which it was retrieved
- field `ctrname` with the name of the register (`EUCTR`, `CTGOV`, `CTGOV2`, `ISRCTN`, `CTIS`) from which that trial was retrieved
- field `record_last_import` with the date and time when that document was last updated using `ctrLoadQueryIntoDb()`
- only for `CTGOV2`: object `history` with a historic version of the trial and with `history_version`, which contains the fields `version_number` (starting from 1) and `version_date`
- all original fields as provided by the register for that trial (see examples [below](https://rfhb.github.io/ctrdata/index.html#trial-records-json-in-databases))
For visualising the data structure for a trial, see this
[vignette section](https://rfhb.github.io/ctrdata/articles/ctrdata_summarise.html#analysing-nested-fields-such-as-trial-results).
## Vignettes
- [Install R package ctrdata](https://rfhb.github.io/ctrdata/articles/ctrdata_install.html)
- [Retrieve clinical trial information](https://rfhb.github.io/ctrdata/articles/ctrdata_retrieve.html)
- [Summarise and analyse clinical trial information](https://rfhb.github.io/ctrdata/articles/ctrdata_summarise.html)
## Example workflow
The aim is to download protocol-related trial information and tabulate the trials' status of conduct.
* Attach package `ctrdata`:
```{r load_ctrdata}
library(ctrdata)
```
* See help to get started with `ctrdata`:
```{r help_package}
help("ctrdata")
```
* Information on trial registers and how they can be used with `ctrdata` (last updated 2024-06-23):
```{r help_registers}
help("ctrdata-registers")
```
* Open registers' advanced search pages in browser:
```{r open_searchpages}
ctrOpenSearchPagesInBrowser()
# Please review and respect register copyrights:
ctrOpenSearchPagesInBrowser(copyright = TRUE)
```
* Adjust search parameters and execute search in browser
* When trials of interest are listed in browser, _copy the address from the browser's address bar to the clipboard_ (you can automate this, see [here](#2-script-to-automatically-copy-users-query-from-web-browser))
* Search used in this example: https://www.clinicaltrialsregister.eu/ctr-search/search?query=cancer&age=under-18&phase=phase-one&status=completed
* Get address from clipboard:
```{r get_clipboard}
q <- ctrGetQueryUrl()
# * Using clipboard content as register query URL:
# https://www.clinicaltrialsregister.eu/ctr-search/search?query=cancer&
# age=under-18&phase=phase-one&status=completed
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one&status=completed
q
# query-term query-register
# 1 query=cancer&age=under-18&phase=phase-one&status=completed EUCTR
```
`r emoji::emoji("bell")` Queries in the trial registers can automatically copied to the clipboard (including for "CTIS", where the URL does not show the query) using our solution [here](#3-script-to-automatically-copy-users-query-from-web-browser).
* Retrieve protocol-related information, transform and save to database:
```{r get_url, include=FALSE}
q <- paste0(
"https://www.clinicaltrialsregister.eu/ctr-search/search?", "
query=cancer&age=under-18&phase=phase-one&status=completed")
ctrOpenSearchPagesInBrowser(q)
```
The database collection is specified first, using `nodbi` (see above for how to specify `PostgreSQL`, `RSQlite`, `DuckDB` or `MongoDB` as backend, see section [Databases](#databases-that-can-be-used-with-ctrdata)).
Then, trial information is retrieved and loaded into the collection:
```{r create_db}
# Connect to (or create) an SQLite database
# stored in a file on the local system:
db <- nodbi::src_sqlite(
dbname = "some_database_name.sqlite_file",
collection = "some_collection_name"
)
```
```{r load_euctr}
# Retrieve trials from public register:
ctrLoadQueryIntoDb(
queryterm = q,
euctrresults = TRUE,
con = db
)
# * Found search query from EUCTR:
# query=cancer&age=under-18&phase=phase-one&status=completed
# * Checking trials in EUCTR...
# Retrieved overview, multiple records of 110 trial(s) from 6 page(s) to be downloaded (estimate: 10 MB)
# (1/3) Downloading trials...
# Note: register server cannot compress data, transfer takes longer (estimate: 100 s)
# Download status: 6 done; 0 in progress. Total size: 9.83 Mb (100%)... done!
# (2/3) Converting to NDJSON (estimate: 2 s)...
# (3/3) Importing records into database...
# = Imported or updated 452 records on 110 trial(s)
# * Checking results if available from EUCTR for 110 trials:
# (1/4) Downloading results...
# Download status: 110 done; 0 in progress. Total size: 62.38 Mb (100%)... done!
# Download status: 29 done; 0 in progress. Total size: 116.74 Kb (100%)... done!
# Download status: 29 done; 0 in progress. Total size: 116.74 Kb (100%)... done!
# - extracting results (. = data, F = file[s] and data, x = none):
# F . F F . F . . F . . . F F . . . . . . . . . . . . . . . . . F . . . F . . .
# . . . F . . . F . . . . . . . . . . F . . . . . . . . . . F . . . . . . . . . . . .
# (2/4) Converting to NDJSON (estimate: 9 s)...
# (3/4) Importing results into database (may take some time)...
# (4/4) Results history: not retrieved (euctrresultshistory = FALSE)
# = Imported or updated results for 81 trials
# No history found in expected format.
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 452
```
Under the hood, EUCTR plain text and XML files from EUCTR, CTGOV, ISRCTN are converted using Javascript via `V8` in `R` into `NDJSON`, which is imported into the database collection.
* Analyse
Tabulate the status of trials that are part of an agreed paediatric development program (paediatric investigation plan, PIP). `ctrdata` functions return a data.frame (or a tibble, if package `tibble` is loaded).
```{r analyse_pips}
# Get all records that have values in the fields of interest:
result <- dbGetFieldsIntoDf(
fields = c(
"a7_trial_is_part_of_a_paediatric_investigation_plan",
"p_end_of_trial_status",
"a2_eudract_number"
),
con = db
)
# Find unique (deduplicated) trial identifiers for trials that have more than
# one record, for example for several EU Member States or in several registers:
uniqueids <- dbFindIdsUniqueTrials(con = db)
# Searching for duplicate trials...
# - Getting all trial identifiers (may take some time), 452 found in collection
# - Finding duplicates among registers' and sponsor ids...
# - 342 EUCTR _id were not preferred EU Member State record for 110 trials
# - Keeping 110 / 0 / 0 / 0 / 0 records from EUCTR / CTGOV / CTGOV2 / ISRCTN / CTIS
# = Returning keys (_id) of 110 records in collection "some_collection_name"
# Keep only unique / de-duplicated records:
result <- subset(
result,
subset = `_id` %in% uniqueids
)
# Tabulate the selected clinical trial information:
with(
result,
table(
p_end_of_trial_status,
a7_trial_is_part_of_a_paediatric_investigation_plan
)
)
# a7_trial_is_part_of_a_paediatric_investigation_plan
# p_end_of_trial_status FALSE TRUE
# Completed 52 24
# GB - no longer in EU/EEA 1 1
# Ongoing 0 2
# Prematurely Ended 3 4
# Restarted 0 2
# Temporarily Halted 1 1
# Trial now transitioned 3 2
```
<div id="workflow-ctgov-example"></div>
* Add records from another register (CTGOV2) into the same collection
The new website and API introduced in July 2023 (https://www.clinicaltrials.gov/) is supported by `ctrdata` since mid-2023 and identified in `ctrdata` as `CTGOV2`.
On 2024-06-25, `CTGOV` has retired the classic website and API used by `ctrdata` since 2015. To support users, `ctrdata` automatically translates and redirects queries to the current website. This helps with automatically updating previously loaded queries (`ctrLoadQueryIntoDb(querytoupdate = <n>)`), manually migrating queries and reproducible work on clinical trials information. Going forward, users are recommended to change to use `CTGOV2` queries.
As regards study data, important differences exist between field names and contents of information retrieved using `CTGOV` or `CTGOV2`; see the [schema for study protocols in `CTGOV`](https://cdn.clinicaltrials.gov/documents/xsd/prs/ProtocolRecordSchema.xsd), the [schema for study results](https://cdn.clinicaltrials.gov/documents/xsd/prs/RRSUploadSchema.xsd) and the [Study Data Structure for `CTGOV2`](https://clinicaltrials.gov/data-api/about-api/study-data-structure). For more details, call `help("ctrdata-registers")`. This is one of the reasons why `ctrdata` handles the situation as if these were two different registers and will continue to identify the current API as `register = "CTGOV2"`, to support the analysis stage.
Note that loading trials with `ctrdata` overwrites the previous record with `CTGOV2` data, whether the previous record was retrieved using `CTGOV` or `CTGOV2` queries.
* Search used in this example: https://www.clinicaltrials.gov/search?cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int
```{r load_ctgov2}
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int",
register = "CTGOV2",
con = db
)
# * Appears specific for CTGOV REST API 2.0
# * Found search query from CTGOV2: cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int
# * Checking trials using CTGOV REST API 2.0, found 100 trials
# (1/3) Downloading in 1 batch(es) (max. 1000 trials each; estimate: 10 MB total)
# Download status: 1 done; 0 in progress. Total size: 9.19 Mb (805%)... done!
# (2/3) Converting to NDJSON...
# (3/3) Importing records into database...
# JSON file #: 1 / 1
# = Imported or updated 100 trial(s)
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 100
```
* Using an example from classic CTGOV:
```{r load_ctgov}
# Retrieve trials:
ctrLoadQueryIntoDb(
queryterm = paste0(
"https://classic.clinicaltrials.gov/ct2/results?",
"cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug"),
con = db
)
# Appears specific for CTGOV Classic website
# Since 2024-06-25, the classic CTGOV servers are no longer available.
# Package ctrdata has translated the classic CTGOV query URL from this call
# of function ctrLoadQueryIntoDb(queryterm = ...) into a query URL that works
# with the current CTGOV2. This is printed below and is also part of the return
# value of this function, ctrLoadQueryIntoDb(...)$url. This URL can be used
# with ctrdata functions. Note that the fields and data schema of trials differ
# between CTGOV and CTGOV2.
#
# Replace this URL:
#
# https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug
#
# with this URL:
#
# https://clinicaltrials.gov/search?cond=neuroblastoma&intr=Drug&aggFilters=ages:child,results:with,status:com
#
# * Found search query from CTGOV2: cond=neuroblastoma&intr=Drug&aggFilters=
# ages:child,results:with,status:com
# * Checking trials using CTGOV REST API 2.0, found 65 trials
# (1/3) Downloading in 1 batch(es) (max. 1000 trials each; estimate: 6.5 Mb total)
# Download status: 1 done; 0 in progress. Total size: 7.30 Mb (914%)... done!
# (2/3) Converting to NDJSON...
# (3/3) Importing records into database...
# JSON file #: 1 / 1
# = Imported or updated 65 trial(s)
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 65
```
* Add records from a third register (ISRCTN) into the same collection
Search used in this example: https://www.isrctn.com/search?q=neuroblastoma
```{r load_isrctn}
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "https://www.isrctn.com/search?q=neuroblastoma",
con = db
)
# * Found search query from ISRCTN: q=neuroblastoma
# * Checking trials in ISRCTN...
# Retrieved overview, records of 12 trial(s) are to be downloaded (estimate: 0.2 MB)
# (1/3) Downloading trial file...
# Download status: 1 done; 0 in progress. Total size: 156.09 Kb (100%)... done!
# (2/3) Converting to NDJSON (estimate: 0.07 s)...
# (3/3) Importing records into database...
# = Imported or updated 12 trial(s)
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 12
```
<div id="workflow-ctis-example"></div>
* Add records from a fourth register (CTIS `r emoji::emoji("bell")`) into the same collection
Queries in the CTIS search interface can be automatically copied to the clipboard so that a user can paste them into `queryterm`, see [here](#2-script-to-automatically-copy-users-query-from-web-browser). Subsequent to the relaunch of CTIS on 2024-07-18, there are more than 8,000 trials publicly accessible in CTIS. See [below](#documents-example) for how to download documents from CTIS.
```{r load_ctis}
# See how many trials are in CTIS publicly accessible:
ctrLoadQueryIntoDb(
queryterm = "",
register = "CTIS",
only.count = TRUE
)
# $n
# [1] 6970
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = paste0(
'https://euclinicaltrials.eu/ctis-public/search#',
'searchCriteria={"containAny":"neonate, neonates"}'),
con = db
)
# * Found search query from CTIS: searchCriteria={"containAny":"neonate, neonates"}
# * Checking trials in CTIS...
# (2/4) Downloading and processing trial data... (estimate: 1 Mb)
# Download status: 20 done; 0 in progress. Total size: 818.42 Kb (100%)... done!
# (3/4) Importing records into database...
# (4/4) Updating with additional data: .
# = Imported 20, updated 20 record(s) on 20 trial(s)
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 20
allFields <- dbFindFields(".*", db, sample = TRUE)
# Finding fields in database collection (sampling 5 trial records per register) . . . . . . . .
# Field names cached for this session.
length(allFields[grepl("CTIS", names(allFields))])
# [1] 628
# root field names in CTIS
ctisFields <- allFields[grepl("CTIS", names(allFields))]
ctisFields[!grepl("[.]", ctisFields)]
# CTIS CTIS CTIS
# "ageGroup" "ageRangeSecondary" "authorizedApplication"
# CTIS CTIS CTIS
# "correctiveMeasures" "ctNumber" "ctPublicStatusCode"
# CTIS CTIS CTIS
# "ctrname" "ctStatus" "decisionDate"
# CTIS CTIS CTIS
# "decisionDateOverall" "documents" "events"
# CTIS CTIS CTIS
# "gender" "lastPublicationUpdate" "lastUpdated"
# CTIS CTIS CTIS
# "publishDate" "record_last_import" "results"
# CTIS CTIS CTIS
# "resultsFirstReceived" "shortTitle" "sponsorType"
# CTIS CTIS CTIS
# "startDateEU" "therapeuticAreas" "totalNumberEnrolled"
# CTIS CTIS CTIS
# "trialCountries" "trialPhase" "trialRegion"
# CTIS
# "trialRegionCode"
# use an alternative to dbGetFieldsIntoDf()
allData <- nodbi::docdb_query(
src = db,
key = db$collection,
query = '{"ctrname":"CTIS"}'
)
# names of top-level data items
sort(names(allData))
# [1] "_id" "ageGroup" "ageRangeSecondary"
# [4] "authorizedApplication" "correctiveMeasures" "ctNumber"
# [7] "ctPublicStatusCode" "ctrname" "ctStatus"
# [10] "decisionDate" "decisionDateOverall" "documents"
# [13] "events" "gender" "lastPublicationUpdate"
# [16] "lastUpdated" "publishDate" "record_last_import"
# [19] "results" "resultsFirstReceived" "shortTitle"
# [22] "sponsorType" "startDateEU" "therapeuticAreas"
# [25] "totalNumberEnrolled" "trialCountries" "trialPhase"
# [28] "trialRegion" "trialRegionCode"
# use yet another alternative
oneTrial <- DBI::dbGetQuery(
db$con, paste0(
"SELECT json(json) FROM ", db$collection,
" WHERE jsonb_extract(json, '$.ctrname') == 'CTIS'",
" LIMIT 1;")
)
# display full json tree
# remotes::install_github("hrbrmstr/jsonview")
if (require(jsonview)) json_tree_view(oneTrial[[1]])
# total size of object
format(object.size(allData), "MB")
# [1] "4 Mb"
```
<div id="workflow-cross-trial-example"></div>
* Analysis across trials
Show cumulative start of trials over time.
```{r analyse_across_trials}
# use helper library
library(dplyr)
library(magrittr)
library(tibble)
library(purrr)
library(tidyr)
# get names of all fields / variables in the collaction
length(dbFindFields(".*", con = db))
# [1] 1657
dbFindFields("start.*date|date.*decision", con = db)
# Using cache of fields.
# - Get trial data
result <- dbGetFieldsIntoDf(
fields = c(
"ctrname",
"record_last_import",
# CTGOV2
"protocolSection.statusModule.startDateStruct.date",
"protocolSection.statusModule.overallStatus",
# EUCTR
"n_date_of_competent_authority_decision",
"trialInformation.recruitmentStartDate", # needs above: 'euctrresults = TRUE'
"p_end_of_trial_status",
# ISRCTN
"trialDesign.overallStartDate",
"trialDesign.overallEndDate",
# CTIS
"authorizedPartI.trialDetails.trialInformation.trialDuration.estimatedRecruitmentStartDate",
"ctStatus"
),
con = db
)
# - Deduplicate trials and obtain unique identifiers
# for trials that have records in several registers
# - Calculate trial start date
# - Calculate simple status for ISRCTN
# - Update end of trial status for EUCTR
result %<>%
filter(`_id` %in% dbFindIdsUniqueTrials(con = db)) %>%
rowwise() %>%
mutate(
start = max(c_across(matches("(date.*decision)|(start.*date)")), na.rm = TRUE),
ctStatus = as.character(ctStatus),
isrctnStatus = if_else(
trialDesign.overallEndDate < record_last_import,
"Ongoing", "Completed"),
p_end_of_trial_status = if_else(
is.na(p_end_of_trial_status) & !is.na(n_date_of_competent_authority_decision),
"Ongoing", p_end_of_trial_status)) %>%
ungroup()
# - Merge fields from different registers with re-leveling
statusValues <- list(
"ongoing" = c(
# EUCTR
"Recruiting", "Active", "Ongoing",
"Temporarily Halted", "Restarted",
# CTGOV
"Active, not recruiting", "Enrolling by invitation",
"Not yet recruiting", "ACTIVE_NOT_RECRUITING",
# CTIS
"Ongoing, recruiting", "Ongoing, recruitment ended",
"Ongoing, not yet recruiting", "Authorised, not started",
"2", "3", "4", "5"
),
"completed" = c(
"Completed", "COMPLETED", "Ended", "8"),
"other" = c(
"GB - no longer in EU/EEA", "Trial now transitioned",
"Withdrawn", "Suspended", "No longer available",
"Terminated", "TERMINATED", "Prematurely Ended",
"Under evaluation", "6", "7", "9", "10", "11", "12")
)
result[["state"]] <- dfMergeVariablesRelevel(
df = result,
colnames = c(
"p_end_of_trial_status",
"protocolSection.statusModule.overallStatus",
"ctStatus", "isrctnStatus"
),
levelslist = statusValues
)
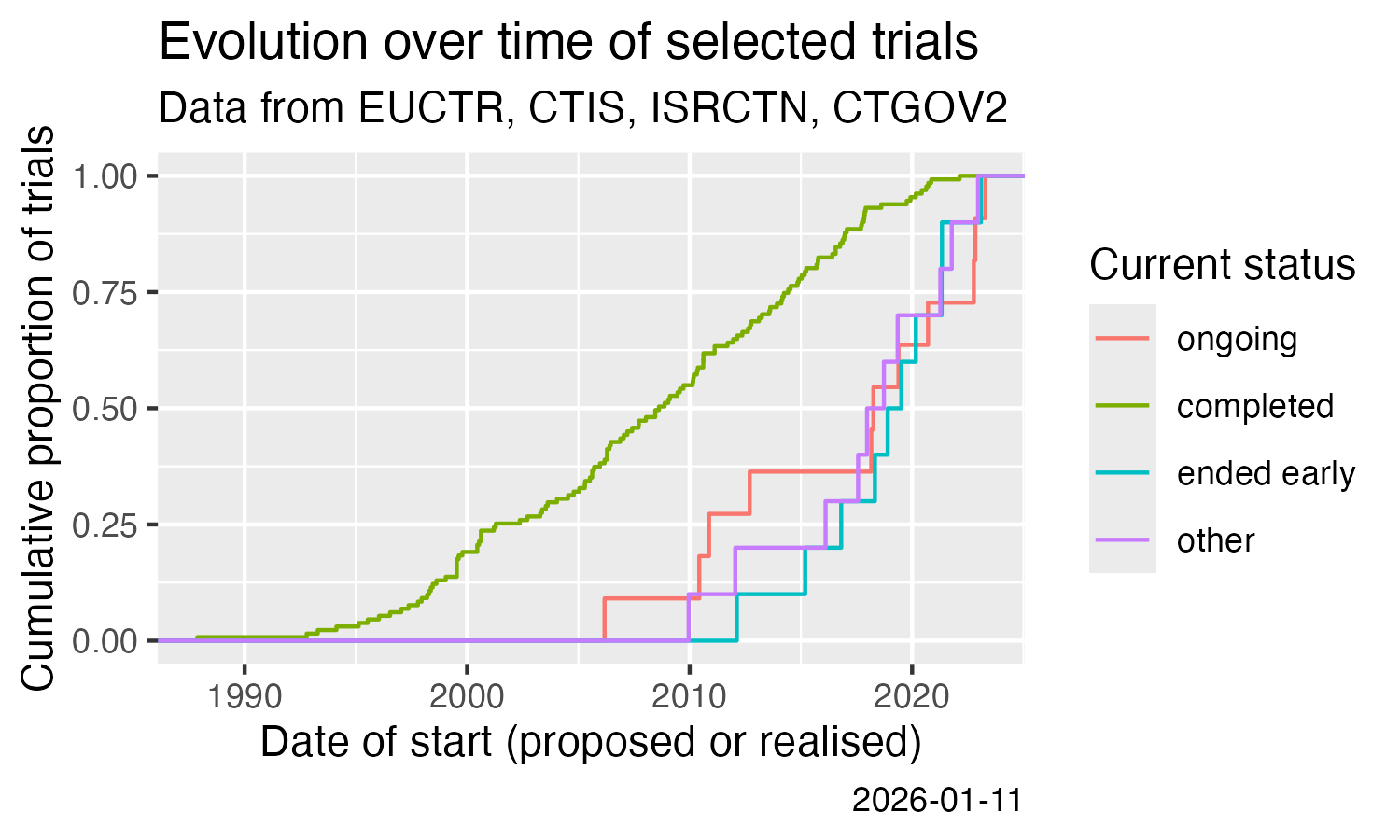
# - Plot example
library(ggplot2)
ggplot(result) +
stat_ecdf(aes(x = start, colour = state)) +
labs(
title = "Evolution over time of a set of trials",
subtitle = "Data from EUCTR, CTIS, ISRCTN, CTGOV2",
x = "Date of start (proposed or realised)",
y = "Cumulative proportion of trials",
colour = "Current status",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_across_registers.png",
width = 5, height = 3, units = "in"
)
```
* Result-related trial information
Analyse some simple result details, here from CTGOV2 (see this [vignette](https://rfhb.github.io/ctrdata/articles/ctrdata_summarise.html) for more examples):
```{r analyse_results}
# Get all records that have values in any of the specified fields:
result <- dbGetFieldsIntoDf(
fields = c(
# fields from CTGOV2 only
"resultsSection.baselineCharacteristicsModule.denoms.counts.value",
"resultsSection.baselineCharacteristicsModule.denoms.units",
"resultsSection.baselineCharacteristicsModule.groups.title",
"protocolSection.armsInterventionsModule.armGroups.type",
"protocolSection.designModule.designInfo.allocation",
"protocolSection.contactsLocationsModule.locations.city",
"protocolSection.conditionsModule.conditions"
),
con = db
)
# Mangle to calculate:
# - which columns with values for group counts are not labelled Total
# - what are the numbers in each of the groups etc.
result %<>%
rowwise() %>%
mutate(
number_of_arms = stringi::stri_count_fixed(
resultsSection.baselineCharacteristicsModule.groups.title, " / "),
is_randomised = case_when(
protocolSection.designModule.designInfo.allocation == "RANDOMIZED" ~ TRUE,
protocolSection.designModule.designInfo.allocation == "NON_RANDOMIZED" ~ FALSE,
number_of_arms == 1L ~ FALSE,
.default = FALSE
),
which_not_total = list(which(strsplit(
resultsSection.baselineCharacteristicsModule.groups.title, " / ")[[1]] != "Total")),
num_sites = length(strsplit(protocolSection.contactsLocationsModule.locations.city, " / ")[[1]]),
num_participants = sum(as.integer(
resultsSection.baselineCharacteristicsModule.denoms.counts.value[which_not_total])),
num_arms_or_groups = max(number_of_arms, length(which_not_total))
)
# Example plot:
library(ggplot2)
ggplot(data = result) +
labs(
title = "Trials including patients with a neuroblastoma",
subtitle = "ClinicalTrials.Gov, trials with results"
) +
geom_point(
mapping = aes(
x = num_sites,
y = num_participants,
size = num_arms_or_groups,
colour = is_randomised
)
) +
scale_x_log10() +
scale_y_log10() +
labs(
x = "Number of sites",
y = "Total number of participants",
colour = "Randomised?",
size = "# Arms / groups",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_results_neuroblastoma.png",
width = 5, height = 3, units = "in"
)
```
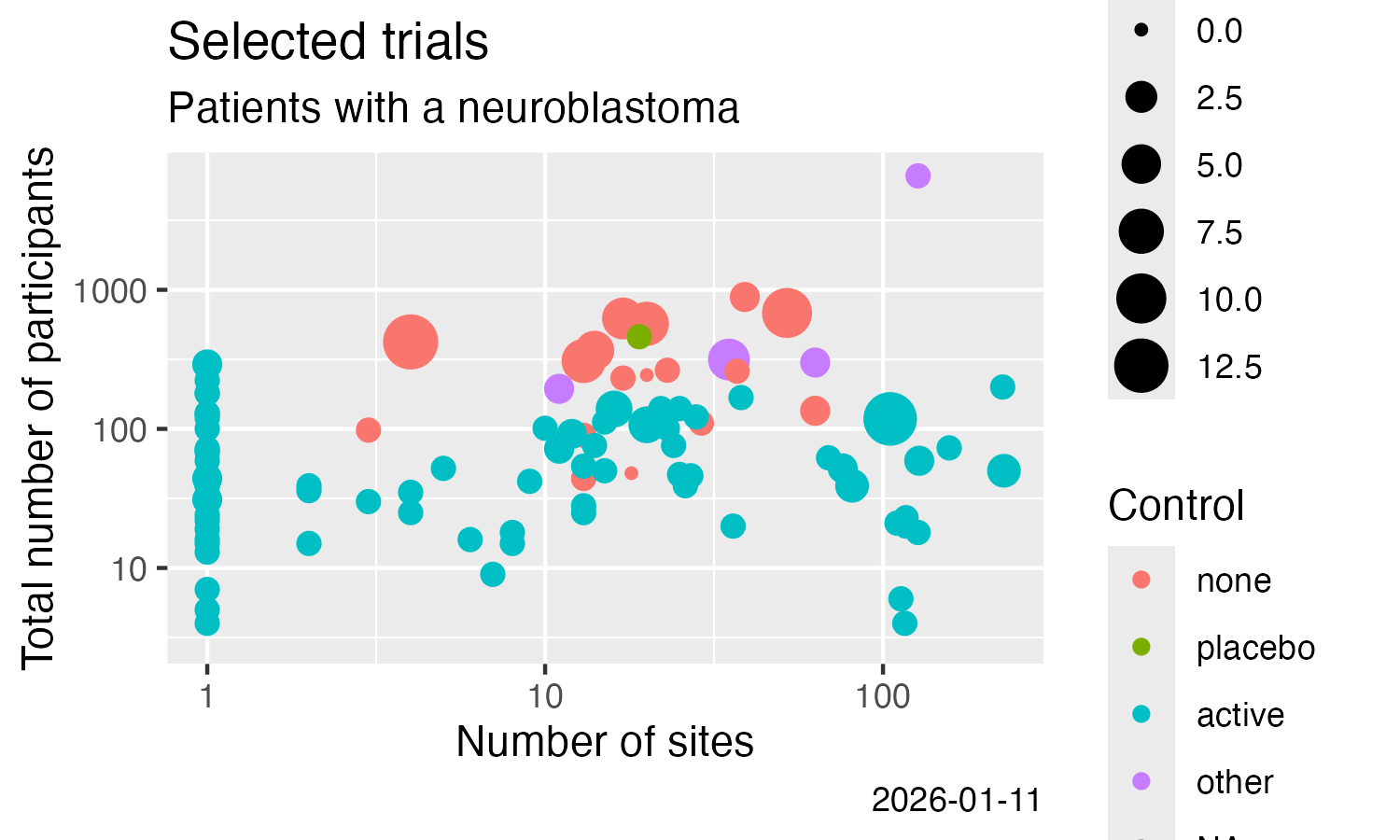
<div id="documents-example"></div>
* Download documents: retrieve protocols, statistical analysis plans and other documents into the local folder `./files-.../`
```{r load_documents}
### EUCTR document files can be downloaded when results are requested
# All files are downloaded and saved (documents.regexp is not used with EUCTR)
ctrLoadQueryIntoDb(
queryterm = "query=cancer&age=under-18&phase=phase-one",
register = "EUCTR",
euctrresults = TRUE,
documents.path = "./files-euctr/",
con = db
)
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one
# [...]
# Created directory ./files-euctr/
# Downloading trials...
# [...]
# = Imported or updated results for 125 trials
# = documents saved in './files-euctr'
### CTGOV files are downloaded, here corresponding to the default of
# documents.regexp = "prot|sample|statist|sap_|p1ar|p2ars|ctalett|lay|^[0-9]+ "
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&type=Intr&recrs=e&phase=1&u_prot=Y&u_sap=Y&u_icf=Y",
register = "CTGOV",
documents.path = "./files-ctgov/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov/
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 35 missing documents
# - Downloading 35 missing documents
# Download status: 35 done; 0 in progress. Total size: 76.67 Mb (100%)... done!
# = Newly saved 35 document(s) for 27 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctgov
### CTGOV2 files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://clinicaltrials.gov/search?cond=neuroblastoma&aggFilters=phase:1,results:with",
documents.path = "./files-ctgov2/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov2/
# - Created directory ./files-ctgov2
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 37 missing documents
# - Downloading 37 missing documents
# Download status: 37 done; 0 in progress. Total size: 77.70 Mb (100%)... done!
# = Newly saved 37 document(s) for 23 trial(s); 0 of such document(s) for 0
# trial(s) already existed in .\files-ctgov2
### ISRCTN files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://www.isrctn.com/search?q=alzheimer",
documents.path = "./files-isrctn/",
con = db
)
# * Found search query from ISRCTN: q=alzheimer
# [...]
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-isrctn/
# - Created directory ./files-isrctn
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 47 missing documents
# - Downloading 29 missing documents
# Download status: 29 done; 0 in progress. Total size: 13.11 Mb (100%)... done!
# Download status: 4 done; 0 in progress. Total size: 13.12 Kb (100%)... done!
# Download status: 4 done; 0 in progress. Total size: 13.12 Kb (100%)... done!
# = Newly saved 25 document(s) for 14 trial(s); 0 of such document(s) for 0 trial(s) already existed in ./files-isrctn
### CTIS files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = paste0(
'https://euclinicaltrials.eu/ctis-public/search#',
'searchCriteria={"containAny":"cancer"}'),
documents.path = "./files-ctis/",
documents.regexp = "sap",
con = db
)
# * Found search query from CTIS: searchCriteria={"containAny":"cancer"}
# * Checking trials in CTIS...
# (1/4) Downloading trial list(s), found 1872 trials
# (2/4) Downloading and processing trial data... (estimate: 100 Mb)
# Download status: 1872 done; 0 in progress. Total size: 167.15 Mb (100%)... done!
# (3/4) Importing records into database...
# (4/4) Updating with additional data: .
# * Checking for documents: . . . . . . . . . . . . . . . . . . .
# - Downloading documents into 'documents.path' = ./files-ctis/
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 16782 missing documents
# - Downloading 4 missing documents
# Download status: 4 done; 0 in progress. Total size: 5.62 Kb (100%)... done!
# Redirecting to CDN...
# Download status: 4 done; 0 in progress. Total size: 3.08 Mb (100%)... done!
# = Newly saved 4 document(s) for 3 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctis
```
```{r cleanup, include=FALSE}
# cleanup
unlink("some_database_name.sqlite_file")
unlink("./files-ctis/", recursive = TRUE)
unlink("./files-euctr/", recursive = TRUE)
unlink("./files-ctgov/", recursive = TRUE)
unlink("./files-ctgov2/", recursive = TRUE)
unlink("./files-isrctn/", recursive = TRUE)
```
## Tests
See also [https://app.codecov.io/gh/rfhb/ctrdata/tree/master/R](https://app.codecov.io/gh/rfhb/ctrdata/tree/master/R)
```{r}
tinytest::test_all()
# test_ctrdata_ctrfindactivesubstance.R 4 tests OK 1.6s
# test_ctrdata_duckdb_ctgov2.R.. 50 tests OK 2.4s
# test_ctrdata_duckdb_ctis.R.... 172 tests OK 15.2s
# test_ctrdata_mongo_local_ctgov.R 51 tests OK 57.7s
# test_ctrdata_other_functions.R 64 tests OK 3.8s
# test_ctrdata_postgres_ctgov2.R 50 tests OK 2.6s
# test_ctrdata_sqlite_ctgov.R... 52 tests OK 56.0s
# test_ctrdata_sqlite_ctgov2.R.. 50 tests OK 2.3s
# test_ctrdata_sqlite_ctis.R.... 194 tests OK 12.5s
# test_ctrdata_sqlite_euctr.R... 105 tests OK 1.3s
# test_ctrdata_sqlite_isrctn.R.. 38 tests OK 21.4s
# test_euctr_error_sample.R..... 8 tests OK 0.9s
# All ok, 838 results (38m 48.8s)
covr::package_coverage(path = ".", type = "tests")
# ctrdata Coverage: 93.68%
# R/zzz.R: 80.95%
# R/ctrRerunQuery.R: 89.16%
# R/ctrLoadQueryIntoDbEuctr.R: 90.03%
# R/utils.R: 90.89%
# R/ctrLoadQueryIntoDbIsrctn.R: 92.11%
# R/dbGetFieldsIntoDf.R: 93.06%
# R/ctrLoadQueryIntoDbCtgov2.R: 94.05%
# R/ctrLoadQueryIntoDb.R: 94.12%
# R/ctrLoadQueryIntoDbCtis.R: 94.13%
# R/ctrLoadQueryIntoDbCtgov.R: 95.04%
# R/dbFindFields.R: 95.24%
# R/ctrGetQueryUrl.R: 96.00%
# R/ctrOpenSearchPagesInBrowser.R: 97.22%
# R/dfMergeVariablesRelevel.R: 97.30%
# R/dfTrials2Long.R: 97.35%
# R/dbFindIdsUniqueTrials.R: 97.77%
# R/dfName2Value.R: 98.61%
# R/ctrFindActiveSubstanceSynonyms.R: 100.00%
# R/dbQueryHistory.R: 100.00%
```
## Future features
* See project outline https://github.com/users/rfhb/projects/1
* Canonical definitions, filters, calculations are in the works (since August 2023) for data mangling and analyses across registers, e.g. to define study population, identify interventional trials, calculate study duration; public collaboration on these canonical scripts will speed up harmonising analyses.
* Merge results-related fields retrieved from different registers, such as corresponding endpoints (work not yet started). The challenge is the incomplete congruency and different structure of data fields.
* Authentication, expected to be required by CTGOV2; specifications not yet known (work not yet started).
* Explore further registers (exploration is continually ongoing; added value, terms and conditions for programmatic access vary; no clear roadmap is established yet).
* ~~Retrieve previous versions of protocol- or results-related information. The challenges include, historic versions can only be retrieved one-by-one, do not include results, or are not in structured format. The functionality available with version 1.17.3 to the extent that is possible at this time, namely for protocol- and results-related information in CTGOV2, only~~
## Acknowledgements
* Data providers and curators of the clinical trial registers. Please review and respect their copyrights and terms and conditions, see `ctrOpenSearchPagesInBrowser(copyright = TRUE)`.
* Package `ctrdata` has been made possible building on the work done for
[R](https://www.r-project.org/),
[clipr](https://cran.r-project.org/package=clipr).
[curl](https://cran.r-project.org/package=curl),
[dplyr](https://cran.r-project.org/package=dplyr),
[duckdb](https://cran.r-project.org/package=duckdb),
[httr](https://cran.r-project.org/package=httr),
[jqr](https://cran.r-project.org/package=jqr),
[jsonlite](https://cran.r-project.org/package=jsonlite),
[lubridate](https://cran.r-project.org/package=lubridate),
[mongolite](https://cran.r-project.org/package=mongolite),
[nodbi](https://cran.r-project.org/package=nodbi),
[RPostgres](https://cran.r-project.org/package=RPostgres),
[RSQLite](https://CRAN.R-project.org/package=RSQLite),
[rvest](https://cran.r-project.org/package=rvest),
[stringi](https://cran.r-project.org/package=stringi) and
[xml2](https://cran.r-project.org/package=xml2).
### Issues and notes
* Please file issues and bugs [here](https://github.com/rfhb/ctrdata/issues). Also check out how to handle some of the closed issues, e.g. on [C stack usage too close to the limit](https://github.com/rfhb/ctrdata/issues/22) and on a [SSL certificate problem: unable to get local issuer certificate](https://github.com/rfhb/ctrdata/issues/19#issuecomment-820127139)
* Information in trial registers may not be fully correct; see for example [this publication on CTGOV](https://doi.org/10.1136/bmj.k1452).
* No attempts were made to harmonise field names between registers (nevertheless, `dfMergeVariablesRelevel()` can be used to merge and map several variables / fields into one).
## Trial records in databases
### SQLite
It is recommended to use nodbi >= 0.10.7.9000 which builds on RSQLite >= 2.3.7.9014 (releases expected in November 2024), because these versions enable file-based imports and thus are much faster:
```{r}
# install latest development versions:
devtools::install_github("ropensci/nodbi")
# requires compilation, for which under MS Windows
# automatically additional R Tools are installed:
devtools::install_github("r-dbi/RSQLite")
```
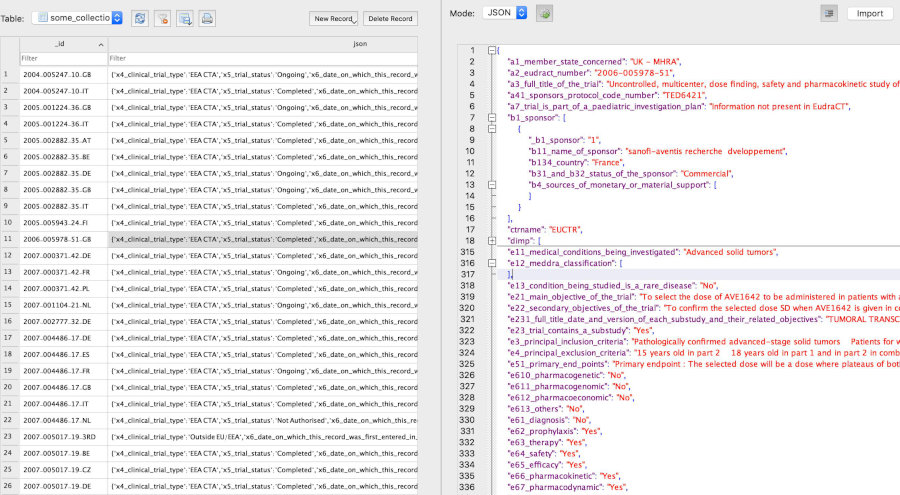
### MongoDB