-
Notifications
You must be signed in to change notification settings - Fork 184
Polymer Glossary
AlexanderSandanov edited this page Apr 14, 2022
·
5 revisions
| Term | Explanation |
|---|---|
| Polymer (strictly this is Polymer molecule ) | A molecule of high relative molecular mass , the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass. _https://goldbook.iupac.org/terms/view/M03667 Basically, the polymer is a molecule that consists of other smaller molecules (that are called monomers) that are united by a special type of chemical bond to form a united structure. |
| Polymerization | The process of converting a monomer mixture of monomers into a polymer. _https://goldbook.iupac.org/terms/view/P04740 |
| Monomer (in all polymer editors, including Ketcher), but truly this is Monomer molecule | A special type of molecule, that in the reactions can form a bond with other monomers and be a building part of the polymer. In Peptide editors, this name is used for the molecules that can be used as components that can be linked by a peptide bond to form the peptide. _A molecule which can undergo polymerization thereby contributing constitutional units to the essential structure of a macromolecule. _https://goldbook.iupac.org/terms/view/M04019 HELM provides the monomer set in *.SDF format here: https://github.com/PistoiaHELM/HELMMonomerSets |
| Peptide | A special type of polymer. Basically, the peptide is a chain of amino acids linked by a special type of bond that is called a peptide bond. Have a very similar structure to PROTEINS, but are much shorter: usually, a peptide has 2-50 monomers, while the protein will contain more. Have a huge biological meaning. _UIPAC definition (very complex, don't read if you are not ready): "Amides __ derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another with formal loss of water. The term is usually applied to structures formed from α-amino acids, but it includes those derived from any amino carboxylic acid." _https://goldbook.iupac.org/terms/view/P04479 Example:  Green and blue parts are amino acids ((L-Valine) and (L-Alanine))The black part is the peptide bond source |
| Polypeptide | Basically, just a not very short peptide:_Peptides __containing ten or more amino acid residues _UIPAC Definition - Source |
| Covalent bond | Chemical link between atoms. Chemically, the covalent bond is created by sharing electron pairs between atoms. In polymer editor, the covalent bond between monomers represents the linkage between compatible connection points within corresponding monomers. Covalent bond canvas representation is reflected by line, color-coded with respect to corresponding R connection point Example 1:  Example 2: Example 2:  IUPAC |
| Hydrogen bond | Hydrogen bonds are created between RNA nucleobase monomers and represented with individual line style (dotted or dashed), with individual color coding. Example: 
|
| Peptide bond | A special type of covalent chemical bond that is able to link amino acids. Consists of 4 atoms (N, H, C, O) that are bond and oriented in a special way:  The dehydration condensation of two amino acids to form a peptide bond (red) with expulsion of water (blue). source This reaction represents beautifully that the Amino group in one acid will join the Carboxyl group in another acid to form the peptide bond. Peptide editors usually represent the peptide bond in a contracted way showing it just as a line linking the monomers and not providing the ability to expand and view the actual structure. Seems that chemists understand what is considered under this type of representation. |
| Connection point, Attachment point | In Peptide editors: part of the amino acids that can contribute its molecules to the peptide bond. Basically, this is a general representation of Amino & Carboxyl groups. Amino acids can have 2 or 3 connections points. Each connection point of the monomer added to the canvas can have 2 states: Free or Occupied |
| Monomer type | In Peptide editors: a group of monomers united by their qualities and connection points properties to ease the navigation and search in the monomer type. Usual types: Peptide (Here are the amino acids and their derivatives), RNA (5 nucleotides + variations of each of their parts), Chem (usually small organic where the structure does not conform to any of the defined polymer types) |
| Complex monomer | Type of RNA monomer that cosist of simple monomer triplet, phophate, shugar and nucleobase. Is used to build simple RNA polymer chains Example: 
|
| Simple RNA monomer | Part of complex RNA monomer. Can be presented as Phosphate, Shugar or Nucleobase |
| Natural monomer | A peptide monomer or simple RNA monomer that can be usually found in nature in living oranisms. In Ketcher polymer editor natural monomers have a single letter ID and can be referenced as parent for unnatural monomers so that while converting polymer notation into sequence Unnatural monomer IDs is replaced by Natural ones. Example: Natural nucleobase type Simple RNA monomer Adenine (ID: A), has a very similar unnatural anlogue 6-ethyladenine (ID: e6A), so that while converting a polymer notation containing e6A into sequence it will be replaced by A |
| Unnatural monomer | Chemically modified natural monomer or fully synthetically created molecule that can be used in polymer design. In Ketcher polymer editor has multi letter ID and can have a reference to corresponding Natural analog. |
| Complimentary nucleobases | Pairs of nucleobase simple RNA monomers that can be connected between each other with hydrogen bond. Adenine (A) is complimentary with Thymine (T) or Uracyl (U) Guanine (G) can be complimentary only with Cytosine (C) |
| Amino acid | Amino acids are the monomers that make up proteins. Each amino acid has the same fundamental structure, which consists of a central carbon atom, also known as the alpha (α) carbon, bonded to an amino group (NH2), a carboxyl group (COOH), and to a hydrogen atom. In the aqueous environment of the cell, both the amino group and the carboxyl group are ionized under physiological conditions, and so have the structures -NH3+and -COO–, respectively. Every amino acid also has another atom or group of atoms bonded to the central atom known as the R group. This R group, or side chain, gives each amino acid protein-specific characteristics, including size, polarity, and pH. https://courses.lumenlearning.com/introchem/chapter/amino-acids/ 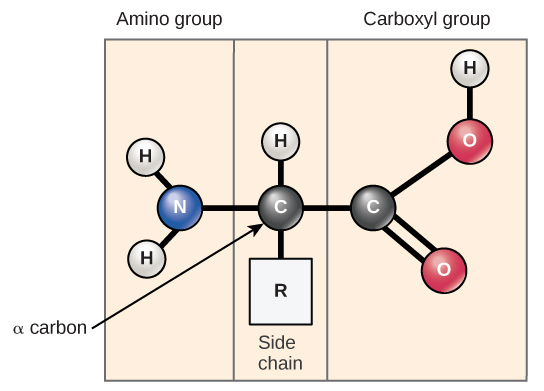 Amino acids can have additional Amino OR Carboxyl group in the R group. In this case, this acid has an additional way to create the peptide bond. List of amino acids from different groups is represented here: https://courses.lumenlearning.com/introchem/chapter/amino-acids/Amino acids have the UIPAC nomenclature https://iupac.qmul.ac.uk/AminoAcid/AA1n2.html |
| Nucleotide | A special class of molecules that serve as monomer units of Nucleic acids. Nucleotide has 3 parts: a nucleobase , a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. In the picture below the Nitrogenous base refers to nucleobase.  Our peptide editor will work with a limited class of Nucleotide. We'll consider only ones with ribose as **sugar ** - they form the RNA nucleic acid. There are several possible nucleobases for RNA monomers the Ketcher peptide editor supports: guanine (G), adenine (A), cytosine (C), thymine (T), uracil (U). Their derivatives can also play the nucleobase role in the RNA polymer. The sugar part of the nucleotide has 3 connection points, phosphate has 2 connection points, nucleobase has 1 connection point. In the Ketcher the Nucleotides will be represented as RNA monomers in the Monomer library. They will have several types: complex type (representing Nucleotide), and simple types (representing Nucleobase, Sugar, Phosphate accordingly). Sugar and Phosphate are called **backbone ** monomers, Nucleobase is considered as **branch **monomers in terms of HELM notation. |
| Chain of Peptide type (or Peptide) in Ketcher peptide editor | Set of monomers linked by the peptide chain with the Amino connection point linked to Carboxyl connection point on each bond and free Amino connection point in the beginning and free Carboxyl connection point on the end of the chain. Will have the number (it will be used when importing to HELM format). Every monomer in the chain should have a number starting from the beginning of the chain to the end of the chain. We consider a single monomer without bonds on the canvas as a chain having the number.
|
| Chain of RNA type in Ketcher peptide editor | Set of RNA monomers linked by the bonds. There is a "true RNA chain" that represents the RNA structure with a backbone consisting of Sugar and Phosphate monomers linked alternately one by one & Nucleobase linking to each Sugar. The backbone starts with Sugar and ends with Phosphate generally. For such a chain, we'll call the beginning of the chain the left monomer in the backbone part and the end of the chain as a monomer on the right end of the backbone part. When other links are created for RNA backbone monomers the resulted chain will be considered RNA chain also, but it will not represent the real RNA structure. Will have the number (it will be used when importing to HELM format). Every monomer in the chain should have a number starting from the beginning of the chain to the end of the chain. We consider a single monomer without bonds on the canvas as a chain having the number. If this monomer is an RNA monomer, then this is an RNA chain and it will be considered as both the beginning and end element of the RNA. |
| Chain of CHEM type in Ketcher peptide editor | Every single monomer of CHEM type that is presented on the canvas. Usually, CHEM monomers are used to represent small organic molecules where the structure does not conform to any of the defined polymer types. It is there to cover drug antibody conjugates and similar situations. CHEM and BLOB are very close in HELM notation, but there is agreement on how they are usually used LINK |
| Peptide monomer in Ketcher peptide editor monomer library | Entity representing the amino acids in peptide editor. Will represent the connection points amino acid has and its structure. |
| HELM File Format | The hierarchical editing language for macromolecules (HELM) is a method of describing complex biological molecules. It is a notation that is machine readable to render the composition and structure of [peptides] (https://en.wikipedia.org/wiki/Peptides), proteins, oligonucleotides, and related small molecule linkers. https://doi.org/10.1021%2Fci3001925 HELM notation is described here: https://pistoiaalliance.atlassian.net/wiki/spaces/PUB/pages/13795362/HELM+Notation |
- Polymer Editor - Glossary
- Polymer Editor - Business Rules
- Polymer Editor - HELM Mapping Rules
-
Polymer Editor - HELM RNA monomers numeration
-
Polymer Editor - Monomer Representation
-
Polymer Editor - Long Chain Polymer Representation
- Polymer Editor Save file
-
Polymer Editor Open file
- Monomer Library - Peptide Monomers
- Monomer Library - RNA Monomers
-
Monomer Library - Chem Monomers
- Monomer Peptide - Adding to the canvas
-
Monomer Peptide - Adding bonds between Peptide monomers
-
RNA Structure - Adding to the canvas
-
RNA Structure - Adding bonds between RNA monomers
-
RNA Structure - Adding hydrogen bonds between RNA monomers
- Chem Structure - Adding to the canvas
-
Chem Structure - Adding bonds between CHEM monomers
- Complex Polymers - Adding bonds between monomers of different types