This is a temporary project (during March 2023?) to host the files for the julia study group (in the context of the Mopo project in preparation for WP4). The project is forked from 'From Python To Julia' to get started but will be expanded dynamically as needed by the members of the study group. The Julia Study Group is a playground to experiment with the julia programming language. There will not necessarily be a big end goal or structure.
Some of the topics that will be addressed:
- basics: dictionaries, arrays, loops, functions
- documentation and modules
- reading/writing files
- using JuMP
- using dataframes
- two step iterative process including the JuMP model
- calling a simple SpineOpt model from within Julia
warning: the links in this readme file are here to help you. However, there is always a risk with clicking on links. So, always check a link before you click on it.
To join the study group, first join the gitter room (button on top of this readme file) and ask there. If you are part of the Mopo project, We'll add you as soon as possible to the Teams meeting and as a developer on github. If you are not of the Mopo project, I guess you are still welcome but you will have to provide a small motivation and you'll have to accept that Mopo project members are prioritised (especially during the online meetings). We'll just have to see what works and what not.
There are two parts to our learning approach. As we start at the very start we will fiddle around with some very basic code. That will be done in the jupyter notebook file and will serve later on as some sort of code library.
Then we will get more serious and we will make a more complex program. We will make a double 'optimisation' loop. The inner loop will be a mixed integer linear formulation. The outer loop will be a monte carlo analysis of the parameters of that formulation. We will also plot the results in some nice figures.
We will do that step by step. The main.jl file will hold the structure (including documentation) and the mod_name.jl file will be a file for each of us where we can try our own version of the algorithm. As we will use the same structure, we will be able to compare our files (in speed and flexibility) and learn from our differences.
During the meetings we can work collaboratively on jupyter lab. Typically that means that we need a server or use a jupyter collaboration programs. We have settled on Binder. The process is the following: The meeting organiser (probably Tars) needs to click the binder button above. You don't have to do that; that would create another instance and you still would not be able to work together. Instead, the organiser will send you the share link during the meeting such that everyone joins the same instance of the jupyter notebook.
For this study group we will obviously use Julia. When I'm learning to code or when I'm trying something out, I typically find a jupyter notebook quite useful. So we'll try that as well.
We will communicate through gitter as this is also used in WP4. Once you click on the button you are invited to the room but you actually still need an application to get it running. There will be some guidance but you might need some extra help.
Gitter has moved to matrix so you need an application that runs matrix. Element is a very solid option but there are other cute ones like Fluffy chat.
Matrix allows for different server domains. By default it selects Matrix.org but in this case we actually want gitter.im so make sure to change to that.
Finally you can make your account with a github account.
If the room is not automatically added, you can just click the gitter button above again.
Note that Matrix is a more secure application and from time to time will ask for verification. Suppose that you change your application from element to fluffychat. It is recommended that you first install and setup fluffychat before you remove element as you might require the old application to verify the new application. Perhaps it is not a problem for public rooms but it might be for your personal chats.
You might not have Julia already so make sure it is installed: https://julialang.org/downloads/
You can check whether it is installed correctly by going to your terminal and type: julia --version (you might have to add julia to the environment path)
You will need some additional Julia packages: https://docs.julialang.org/en/v1/stdlib/Pkg/
In short, in the terminal type: julia to open the julia environment and type ] to get into the package manager (the text will typically become blue). To add a package type add with the package name, e.g. add IJulia. (Afterwards, you can then exit the package manager with backspace and exit Julia by typing: exit())
A short list of packages we will probably use:
- IJulia (to make julia available as a kernel in the jupyter notebook)
- Colors
- Plots
- JuMP (for mixed integer linear formulations such as in SpineOpt)
- GLPK (to use the glpk solver in JuMP; you can also use other JuMP compatible solvers)
- SpineOpt (we'll install the developer version together in one of the meetings)
To install and use Jupyter, you need to install Python: https://www.python.org/downloads/. The package manager for python is pip (there is also the option with a conda environment but I know nothing about that). Again you can check whether python is installed correclty with the command python --version.
You can install Jupyter through pip (from python): pip install jupyterlab. (Be sure to install jupyterlab and not just jupyter; jupyterlab is the successor of jupyter and jupyter does not include jupyterlab by default.)
To open a jupyter notebook you can open jupyterlab with the commandline jupyter lab (the python scripts folder needs to be in your environment variable PATHS) or use your own preferred application. My personal favorite:
vs code.
Feel free to add training materials that you found useful.
From Python To Julia: The original project which is dedicated to the differences between Python and Julia and remains a separate project.
Advanced Julia training: A project by the ESIM group at the KU Leuven to get students or collegues up to speed with Julia. The owner of the repository told me that if there were any issues or suggestions, you can contact them through the foreseen issue list. They'll try to respond as soon as possible.
JuMP: The mixed integer linear model formulation package for Julia.
JuMP compatible solvers: A list and package names of solvers that can be used in JuMP
DataFrames: To get started with dataframes I started from a tutorial on dataframes in jupyter notebooks. It goes pretty deep but I only played around in the first two to get the basics, then I used the search function in the official documentation when I still needed something else.
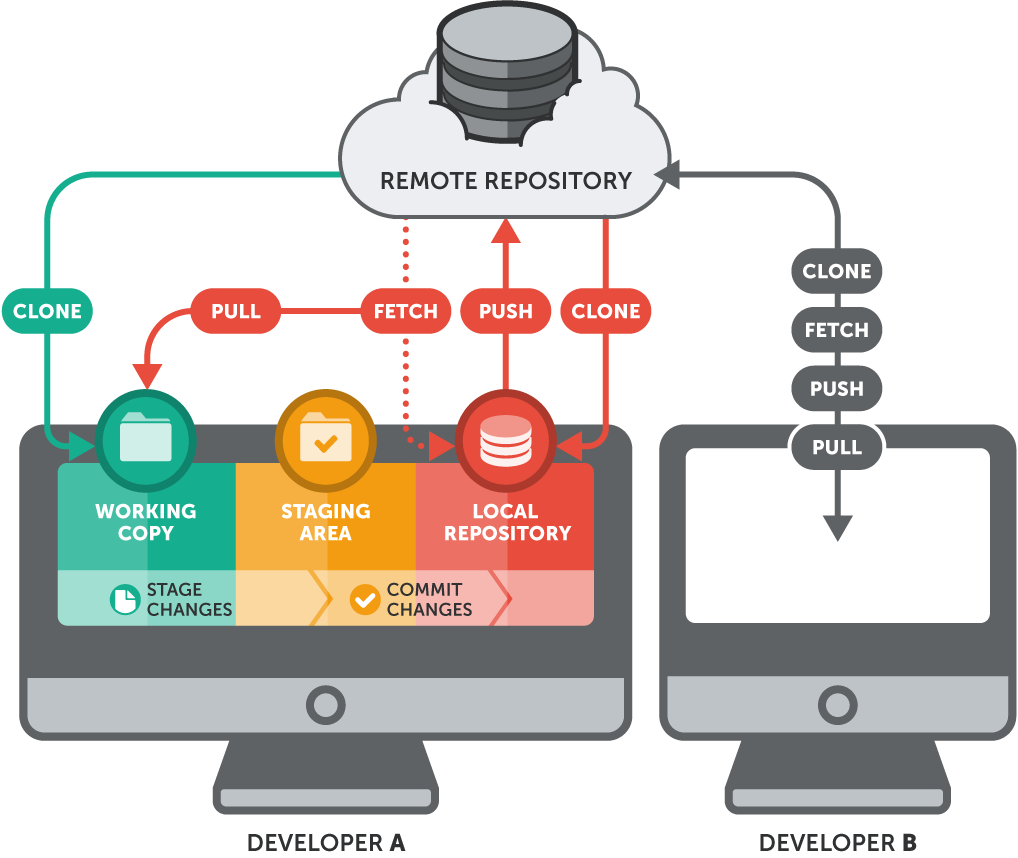
git-tower also explains git quite nicely. Below is an overview of the principle.
The meetings are scheduled to be held every friday from 10:00 to 12:00 (GMT) in Teams. The first hour follows the schedule below, the second hour is a more relaxed continuation of the discussion. The meetings will mostly be used to share our experiences. The agenda is a guideline; additional topics are certainly welcome. Further discussions are also possible in the gitter rooms.
day/month/2023
03/03
homework:
- install everything as indicated above
- read juliastudygroup.ipynb
- add basic julia stuff that you encountered and that you found interesting or useful
agenda:
- if there are newcomers: brief overview of last meeting (by means of this readme)
- brief discussion on any issues during installation
- playing around with the jupyter notebook to exchange our understanding of the basics of Julia
10/03
homework:
- someone should check the code for working with a main file and modules
- add your own mod_name to the repository
- write input and output files (json, excel, csv, ...) without and with DataFrames
agenda:
- discuss io-file
- discuss DataFrames
17/03
homework:
- someone should create some code for benchmarking
- create a linear model with JuMP, you can make it as complex as you like
- bonus: use the loading and saving of files to store your parameters and results
agenda:
- explanation of the benchmarking
- discussion and comparison of linear models with JuMP
24/03
homework:
- create a sampling method for the parameters in the linear model, at least monte carlo but you can do something more interesting as well
- bonus: use parallel computing for the different samples
agenda:
- discuss and compare sampling methods
- link random sampling and linear model with parameteranalysis
31/03
homework:
- visualise the results
agenda:
- discussion and comparison of the visualisation techniques
- preparation of the next session: install the developer version of spineopt
07/04
homework:
- Play around with calling SpineOpt through Julia (create database, pass it to SpineOpt and read the results). Your code does not need to run. In other words, the idea is to get questions and it is not necessary to already have the answers.
- bonus: replace your jump model with the SpineOpt model in your code
resources:
- It is recommended to install the developer version of SpineOpt and SpineInterface. Besides the intallation instructions you also get information for the external monthly user meeting where you can get some additional support. There should be a meeting on 04/04 at 14:00 (UCT).
- for creating a database you can get started with this example
agenda:
- discussion on SpineOpt in Julia as a preparation for the deepdive event
11/04-12/04 SpineOpt deepdive event