-
Notifications
You must be signed in to change notification settings - Fork 0
Architecture
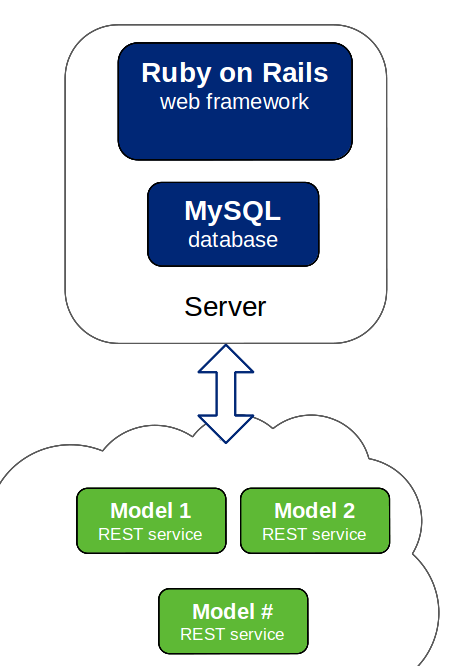
BGFit is developed using open-source frameworks and free libraries allowing for a high dregree of flexibility and creating a modular system. Ruby on Rails, MySQL, Octave, MathJax and Google Chart Tools are some examples.
The application is designed using a model-view-controller architecture effectively separating data-management and dynamic modeling that is performed using extensions that are decoupled from the web-application.
The modeling extensions only require the implementation of the necessary interface and for it to be deployed on a location that is accessible by BGFit. This approach allows for every component of BGFIt to be deployed online, encouraging collaboration and the reutilization of these tools. Nevertheless, it can also be used in a local installation.
The default modeling extension pack that describes different bacterial growth models (such as Baranyi, Gompertz, Logistics, etc...) is implemented in Octave/Matlab. These modeling extensions are also released with BGFit and are described in their documentation, as well as a template model from which all the implemented models derive. This provides a starting point for users to create their own models.
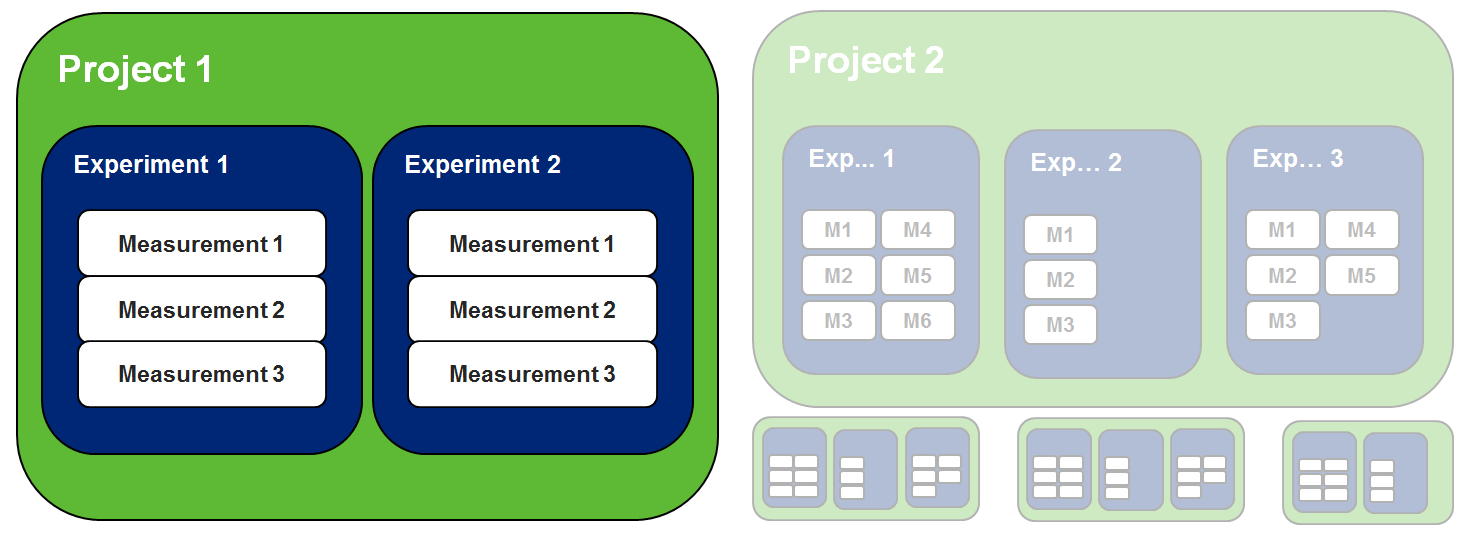
Data management is divided in three layers: project (top-level folder) -> experiment (folder) -> measurement (actual data)
- Project: Top-level folder where permissions and other properties are set
- Experiment: Folder to organize and aggregate data by typology
- Measurement: Actual data